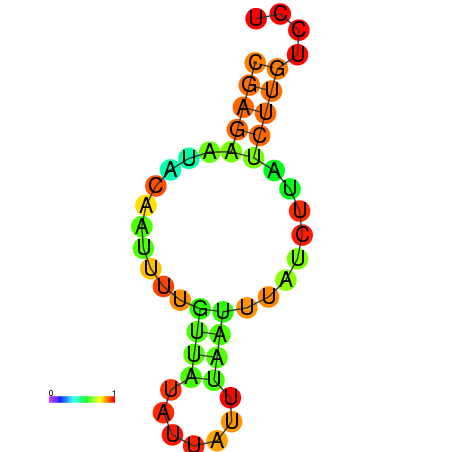
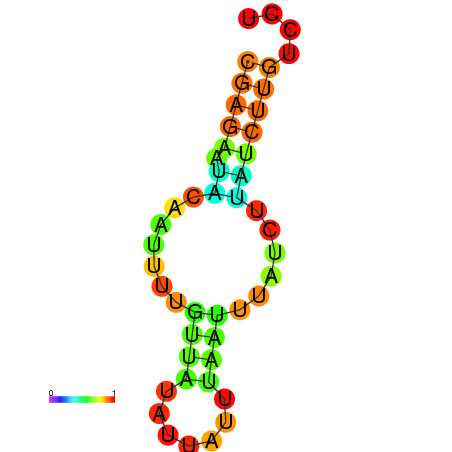
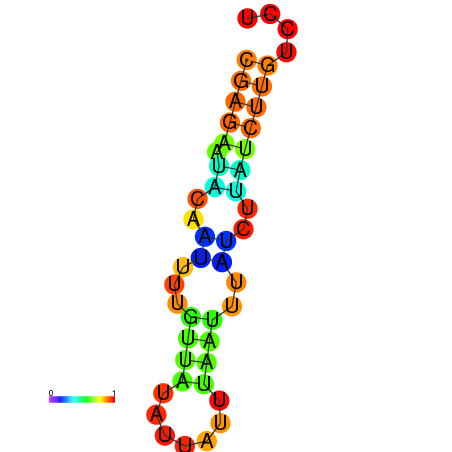
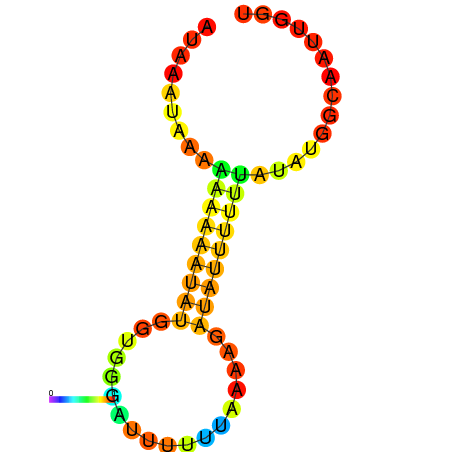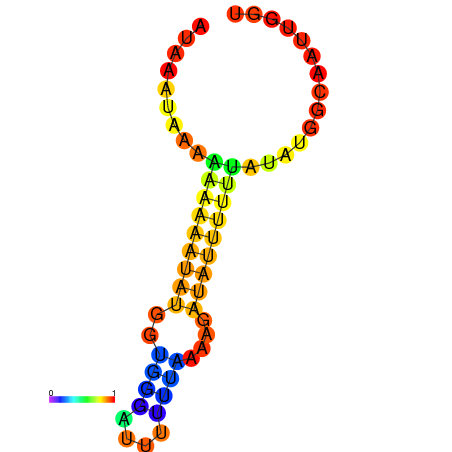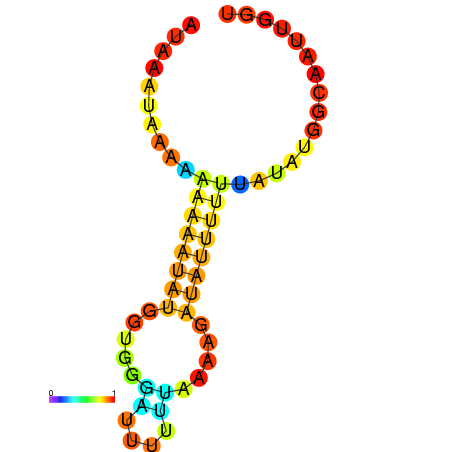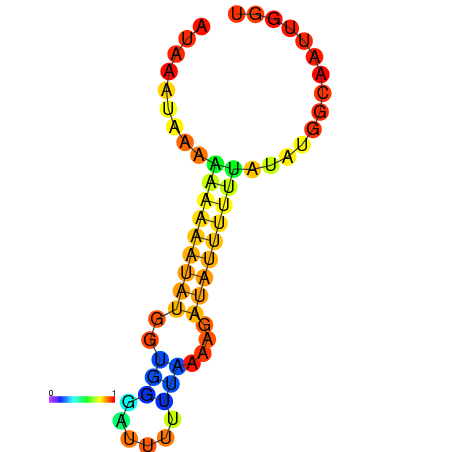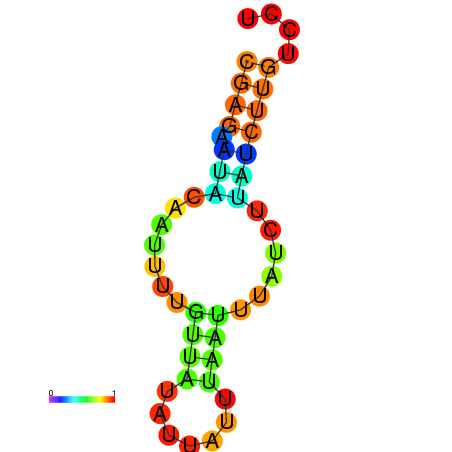| dm3 |
chr2RHet:1322818-1322960 - |
TCCGATGACCCATGGCG----G--GAGAGGATTATGAAAGACAATTTGAAATTAAATATCTGTGT-GTGAAATTATT--------------TAAAAGATTAATTTCACGCGAAGATATTTAT----TTTTACATTTGTCCCTATAATAACCGGGGT----------------------------------------G---------------CCA---ACATACAT |
| droSim1 |
chr2R:265657-265782 + |
AGGC------------------A--------------------ATTTTCAATTAAATATCTGTGT-GTGGAATTATTAAAAACAATGAGTTCAAAGGTTTAGTTTCACGTAAAGATATTTATTTTG-TTTAAATTT-TCCCTAGAATAACCAGTAT----------------------------------------G---------------CCG---ACATAAA- |
| droSec1 |
super_55:122657-122779 + |
AAGT------------------A-------------------AACTTAAAATTATAAATATGTGT-GTGAAATTAATAAA--CGGTAAGTTCGAAAGTTTAGTTTCACGCGAAAGTATTTAT----TTTTACATTTGTCCATAGAATAACCGGGGG----------------------------------------G---------------CCA---ACATAAA- |
| droYak2 |
chr2h_random:1837227-1837349 - |
AAGA------------------C-------------------AACTTAAATTTTTATATCTGTGT-GGGAAATTAAGAAA--CAGTGAGTTCGAAAATTTAGTTTCGTGTAAAGTTATTTAT----TTCTGCATTTGTCCCTAGAATAACCGAGGT----------------------------------------C---------------CCA---ACATAAA- |
| droEre2 |
scaffold_4845:21278391-21278511 + |
CTCCATAACTCAACTT-ATCTT--AATCGGATTTTGAAGTGGCA---TACCT-CTAAATGTTTGTGGTGCAATTATC--------------TAAAAAACTGCATTCAAATTTAAAAATTTATTTTT-TTTACATTTTTTTGTA----------------------------------------------------------------------------------- |
| droAna3 |
scaffold_13334:5190-5293 + |
AAAA------------------AA-------TTAGTAAAAAAAATAATAAATAAAAAAAAATATG-GTGGGATTTTT--------------TAAAAGATATTTTTTATATGGCAATTGGT----------------------------------------------------------------AGTTAAATATTAG---------------TCT---TTTTAAGA |
| dp4 |
Unknown_group_37:8363-8450 + |
GGCAATGAGG--------------------------------------------------------------------------------TAGGAGGGACAATTTTGTAT-----------------------------TCTATAATTATCGTTAGACCTACGCAAAGGATACATTTTACTGCGGG----------T---------------TGT---ATATTTT- |
| droPer1 |
super_5473:1216-1318 + |
TATA------------------C----------------------------------GTGTGTAA-GTGGAACTATG--------------CAGGGGAATGATTTCACATGAAGAAAGTCGATTCG-CCTGTACC------TATTGCATTCC-TA-----------------------------AGCTGAGGTTT-A---------------AGC---TTTAACAT |
| droWil1 |
scaffold_180772:5217896-5218034 + |
GCCGTTGACCAGTGGCG----G--CG----------------CCAAAGTGC--ACATATTCTTGT-CAGAGTTAGCTGGATAGTATGTTCCTGAAAATTCAATTA-AAATTGAA-----------------------------AGAGGACTTG--------------------------------TTGCAATGAATTTTTTTTTAATATTTGTCTAGATTTCGACC |
| droVir3 |
scaffold_291:1622-1745 - |
AAAG------------------AA-------TCATTAGATAAATCTTTAAAATATAGATTTTTTT-TTACCATATTT--------------GTAATGATTTTTGGATTGCGAAATTTTTTAT----TTAAATTGTTAATTATATAATATATTAAAT----------------------------------------A---------------TTT---ATATATAT |
| droMoj3 |
scaffold_6473:7010400-7010528 + |
AAAGACAAAAAAAAAT-AAAAT--AA----------------AAAAAATAAACAAACAAAAGTGT-GTAA---------------AGAGTGCAATGCCAACAAATCATACGTAGATTTCTAT----ATTTACATATGTACATAAATATATATATGT----------------------------------------A---------------TGC---ACATACAT |
| droGri2 |
scaffold_14906:12727535-12727642 + |
TGCC------------------GAGAAGGG------------AAAACGAGAATACAATTTTGTTA-TATTATTTAATTTA--TCTTA-TCTTG------------------------------------TCCTTTTATTTGAAAGATAATTTGA------------------------------ATTTGAATAAATT---------------TAT---ATTTGTAT |