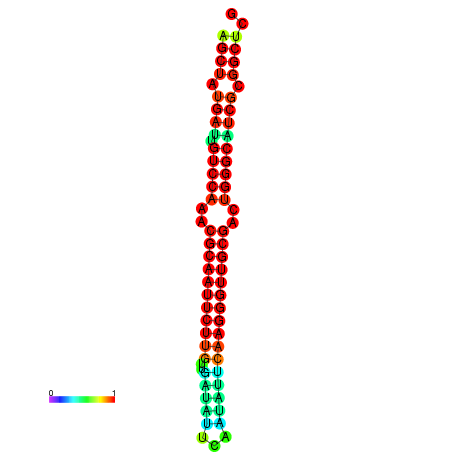
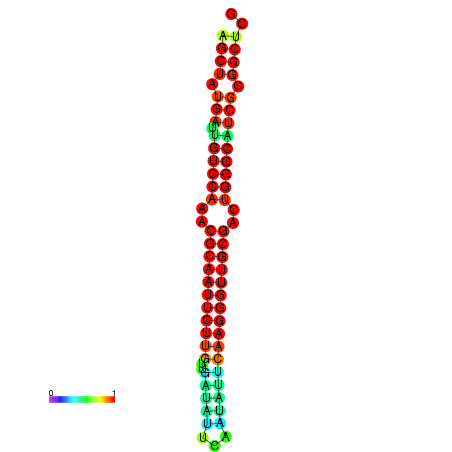
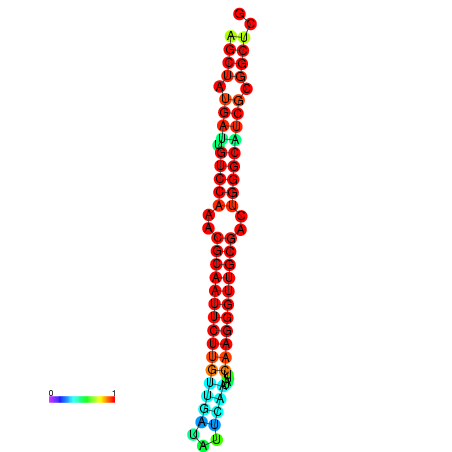
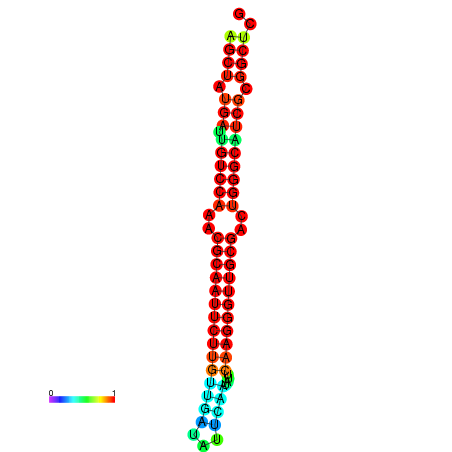

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:17308091-17308220 + | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droSim1 | chr3L:16646167-16646296 + | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droSec1 | super_0:9359464-9359593 + | AAATAAATATTTGGTTAATTCAATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droYak2 | chr3L:16762572-16762701 - | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droEre2 | scaffold_4784:16541302-16541431 - | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droAna3 | scaffold_13337:8265335-8265463 + | CAAAATAT-TTTGGTTAATTCAATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGAAT-TCAAATACTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATACGAATACAACGAAGAGAACCAT--TTC |
| dp4 | chrXR_group6:9338098-9338227 - | ATCAAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TCCAATACTCAAGGGTTGTGACTGGACATCGCGGCTCGAAATAAGAATACAACCCAGCAACCAAA--ATC |
| droPer1 | super_35:387455-387584 + | ATCAAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TCCAATACTCAAGGGTTGTGACTGGACATCGCGGCTCGAAATAAGAATACAACCCATCAACCAAA--ATC |
| droWil1 | scaffold_180955:2086476-2086593 + | A---AAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATT-TCAAATACTCAAGGGTTGCATCTGGGCATCGCGGCTCGAAATAAGAATACA---------CCCAA--TAA |
| droVir3 | scaffold_13049:5991414-5991542 - | GTATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATT-TCAAATACTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATGAGAATCCAACCCA-CAACCAAA--CAA |
| droMoj3 | scaffold_6680:4316890-4317021 - | AAACATATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATTTTTAAATACTCAAGGGTTGCATCTGGGCATCGCGGCTCGAAATGAGAATCCAACACAAA-GCCAAAATATA |
| droGri2 | scaffold_15110:13121815-13121944 + | TAACAAACATTTGGTGAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATT-TCAAAAACTCAAGGGTTGCATCTGGGCATCGCGGCTCGAAATGAGAATACAACACAAAAATCAAA--ACA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:17308091-17308220 + | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droSim1 | chr3L:16646167-16646296 + | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droSec1 | super_0:9359464-9359593 + | AAATAAATATTTGGTTAATTCAATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droYak2 | chr3L:16762572-16762701 - | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droEre2 | scaffold_4784:16541302-16541431 - | AAATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TTCAATATTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATAAGAATACAACTCGGCAACCAAA--TCA |
| droAna3 | scaffold_13337:8265335-8265463 + | CAAAATAT-TTTGGTTAATTCAATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGAAT-TCAAATACTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATACGAATACAACGAAGAGAACCAT--TTC |
| dp4 | chrXR_group6:9338098-9338227 - | ATCAAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TCCAATACTCAAGGGTTGTGACTGGACATCGCGGCTCGAAATAAGAATACAACCCAGCAACCAAA--ATC |
| droPer1 | super_35:387455-387584 + | ATCAAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATA-TCCAATACTCAAGGGTTGTGACTGGACATCGCGGCTCGAAATAAGAATACAACCCATCAACCAAA--ATC |
| droWil1 | scaffold_180955:2086476-2086593 + | A---AAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATT-TCAAATACTCAAGGGTTGCATCTGGGCATCGCGGCTCGAAATAAGAATACA---------CCCAA--TAA |
| droVir3 | scaffold_13049:5991414-5991542 - | GTATAAATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATT-TCAAATACTCAAGGGTTGCGACTGGGCATCGCGGCTCGAAATGAGAATCCAACCCA-CAACCAAA--CAA |
| droMoj3 | scaffold_6680:4316890-4317021 - | AAACATATATTTGGTTAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATTTTTAAATACTCAAGGGTTGCATCTGGGCATCGCGGCTCGAAATGAGAATCCAACACAAA-GCCAAAATATA |
| droGri2 | scaffold_15110:13121815-13121944 + | TAACAAACATTTGGTGAATTCGATTTTTAGCTATGATTGTCCAAACGCAATTCTTGTTGATT-TCAAAAACTCAAGGGTTGCATCTGGGCATCGCGGCTCGAAATGAGAATACAACACAAAAATCAAA--ACA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-29.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-29.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|---|---|
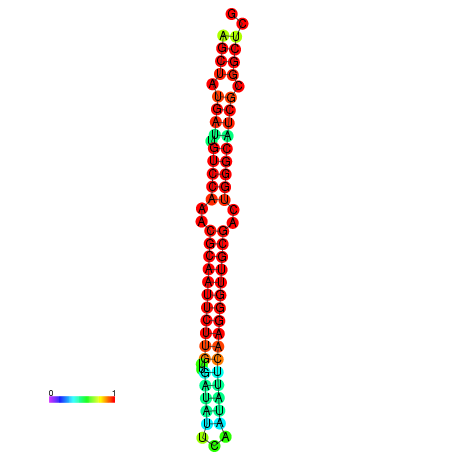 |
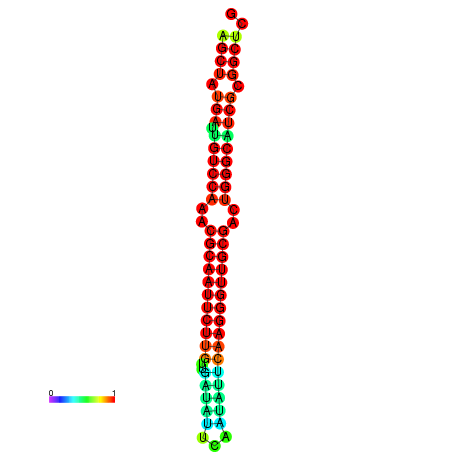 |
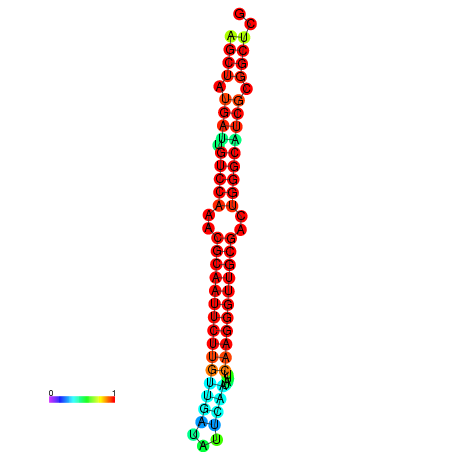 |
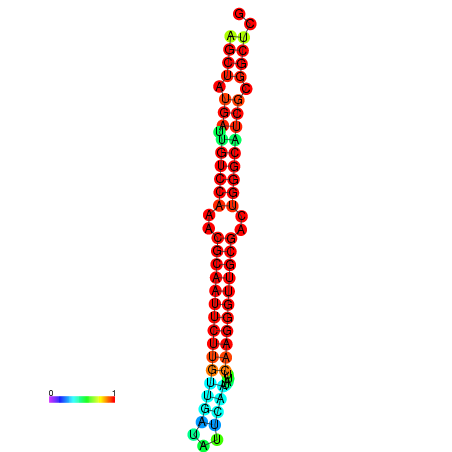 |
 |
| dG=-29.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|---|---|
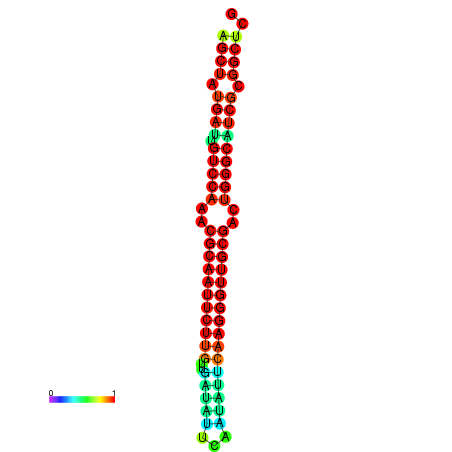 |
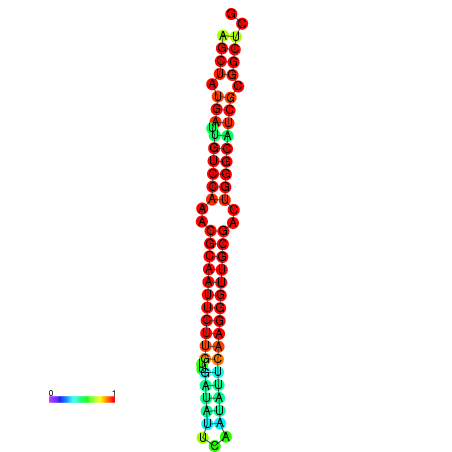 |
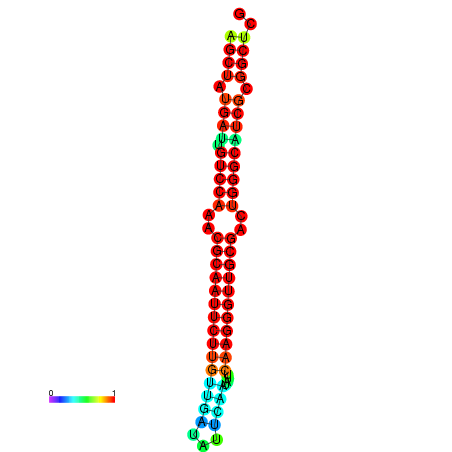 |
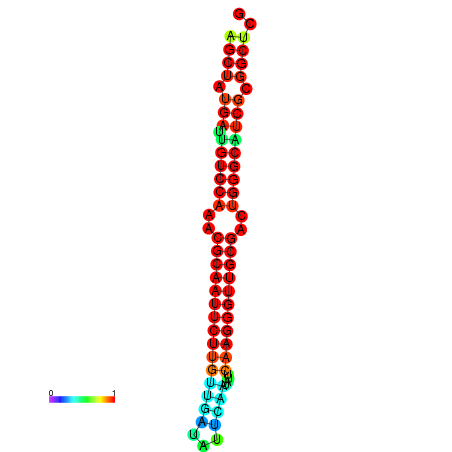 |
 |
| dG=-29.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|---|---|
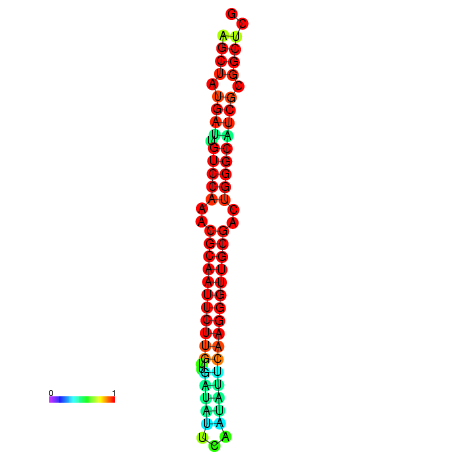 |
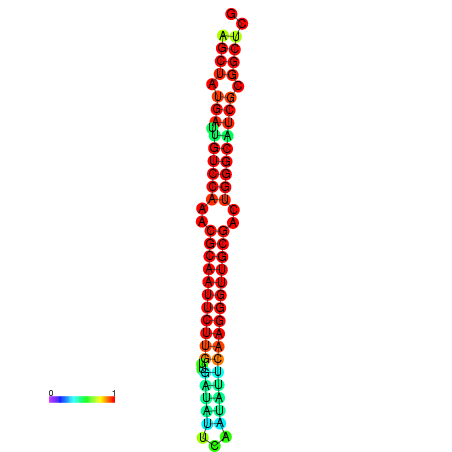 |
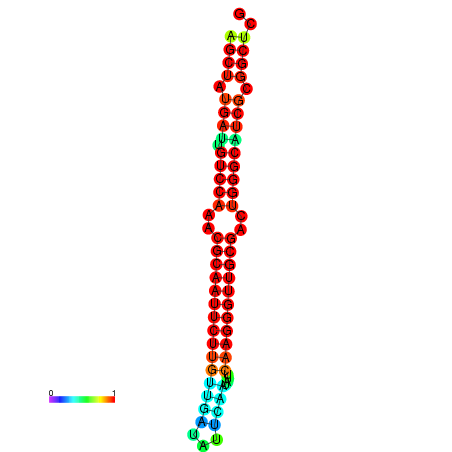 |
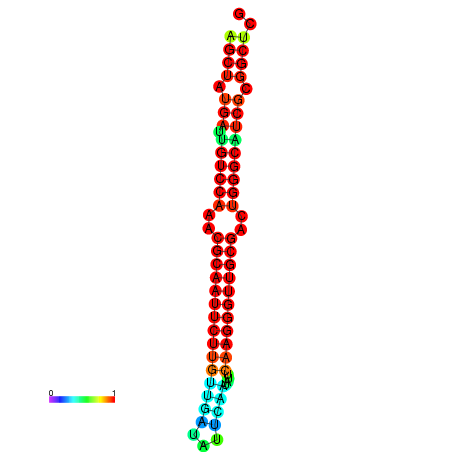 |
 |
| dG=-29.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|---|---|
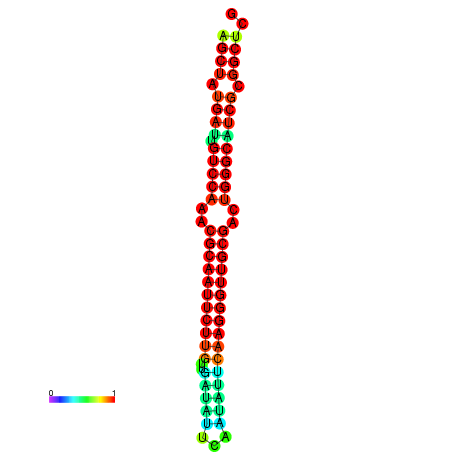 |
 |
 |
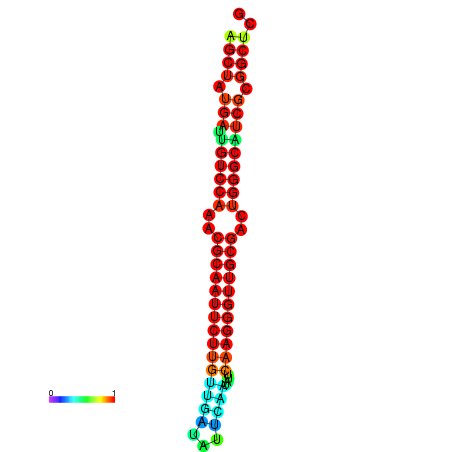 |
 |
| dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-27.7, p-value=0.009901 |
|---|---|---|---|---|
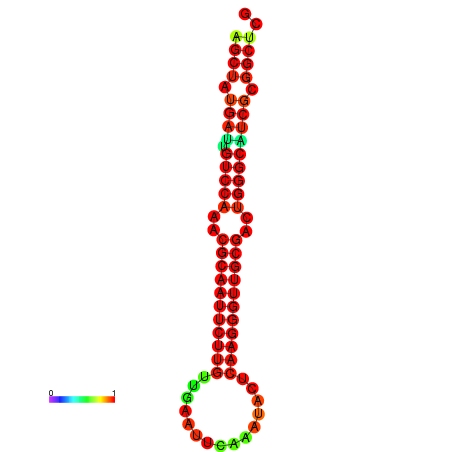 |
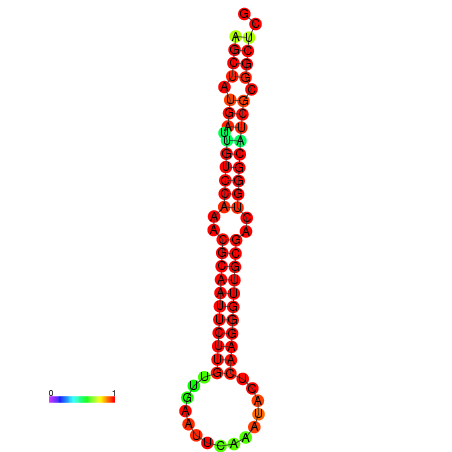 |
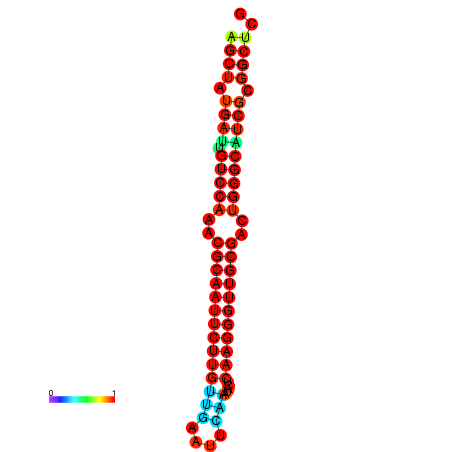 |
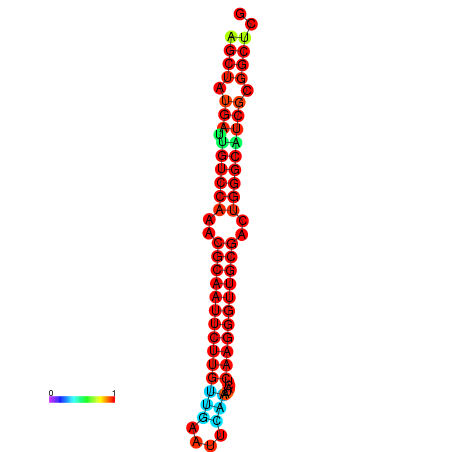 |
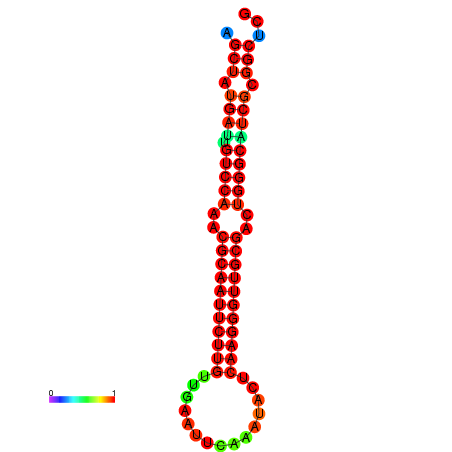 |
| dG=-27.0, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-26.9, p-value=0.009901 |
|---|---|---|---|---|
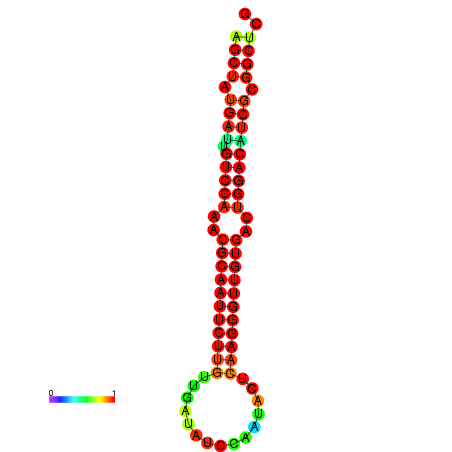 |
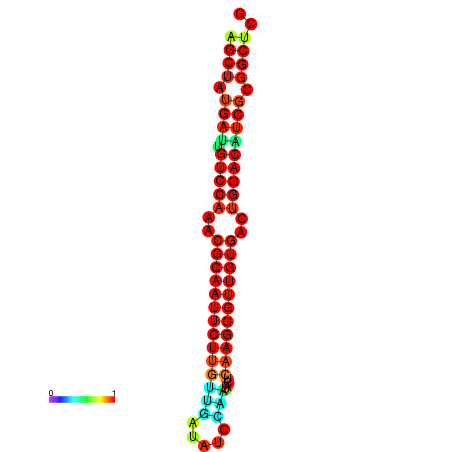 |
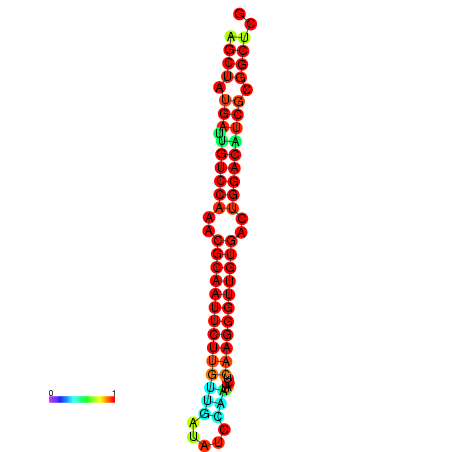 |
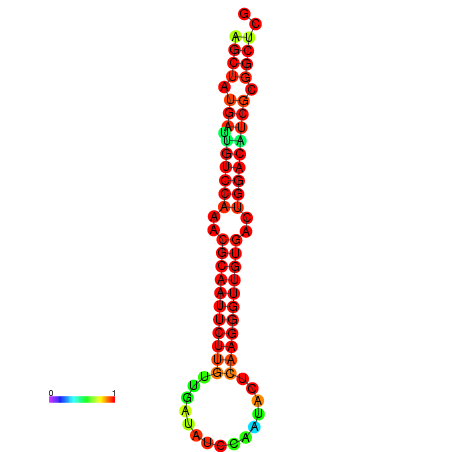 |
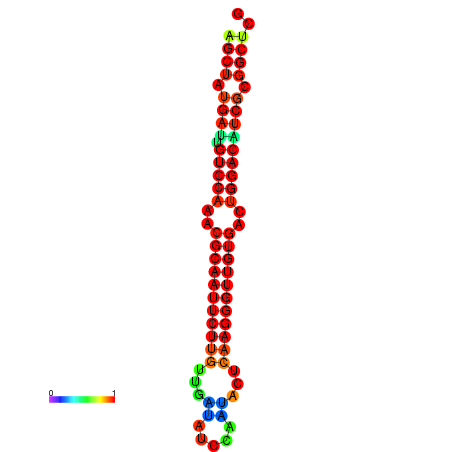 |
| dG=-27.0, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-26.9, p-value=0.009901 |
|---|---|---|---|---|
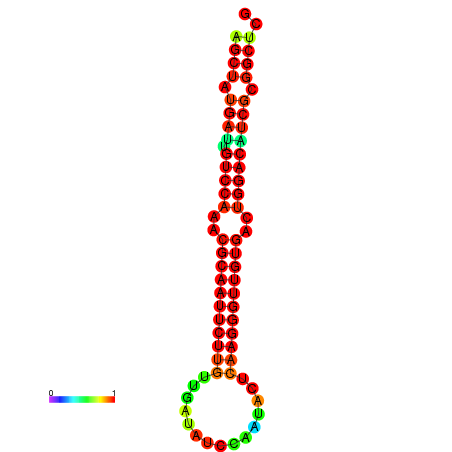 |
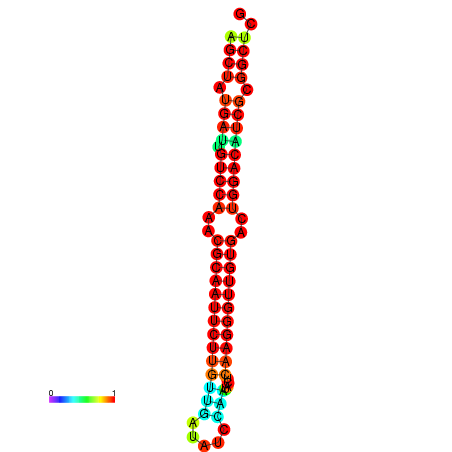 |
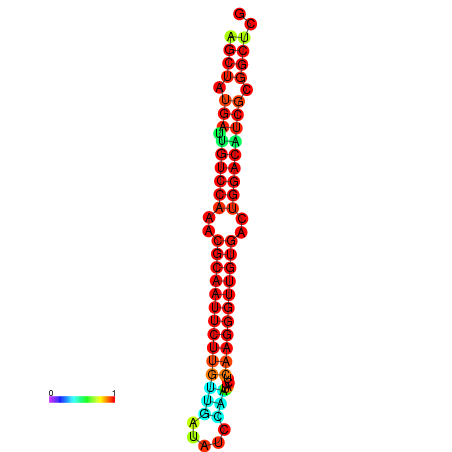 |
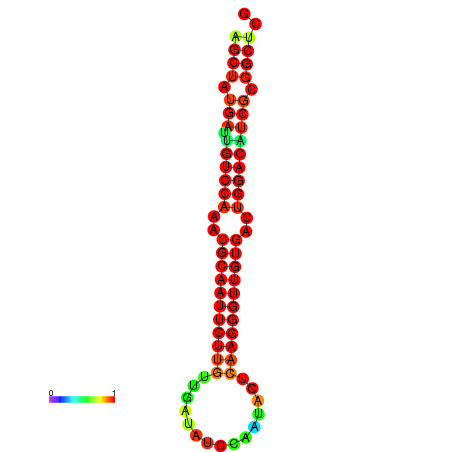 |
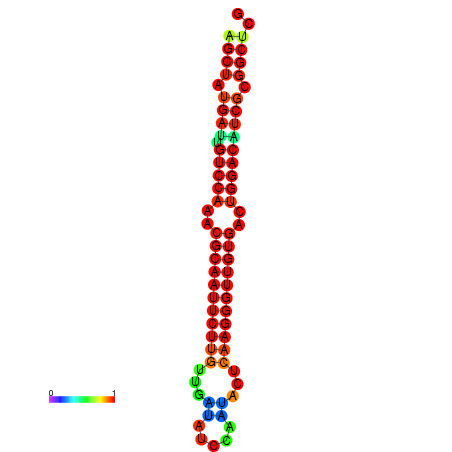 |
| dG=-25.3, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
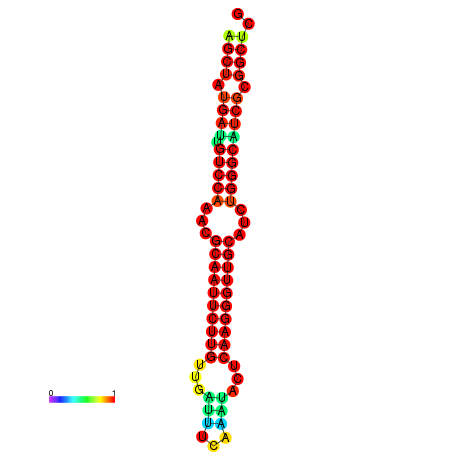 |
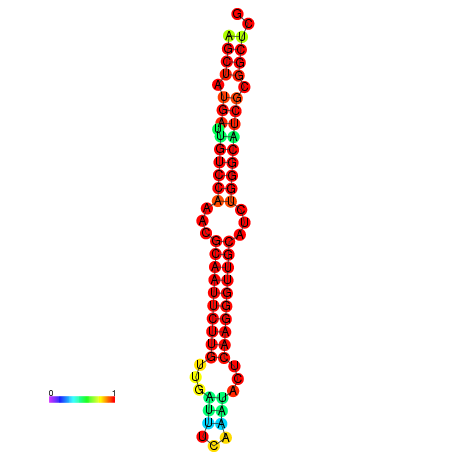 |
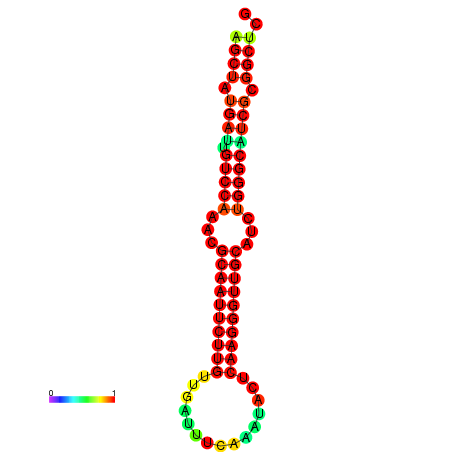 |
 |
 |
| dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
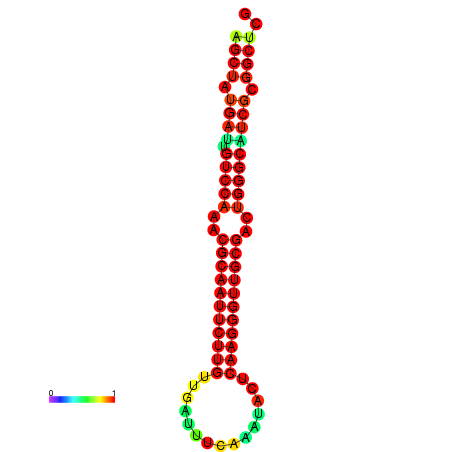 |
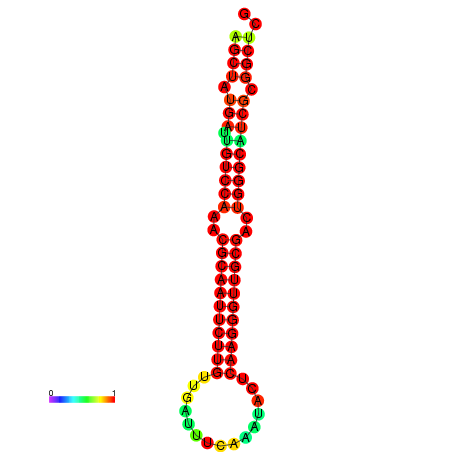 |
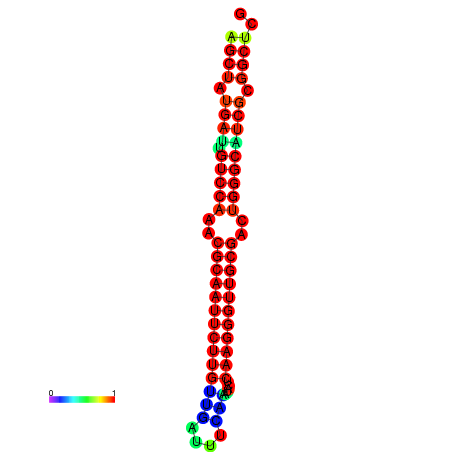 |
| dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-25.1, p-value=0.009901 |
|---|---|---|---|---|
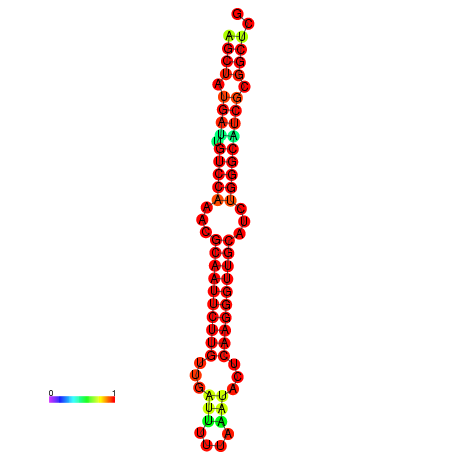 |
 |
 |
 |
 |
| dG=-25.2, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|---|
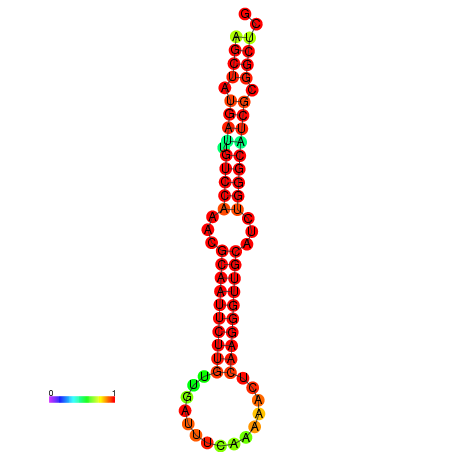 |
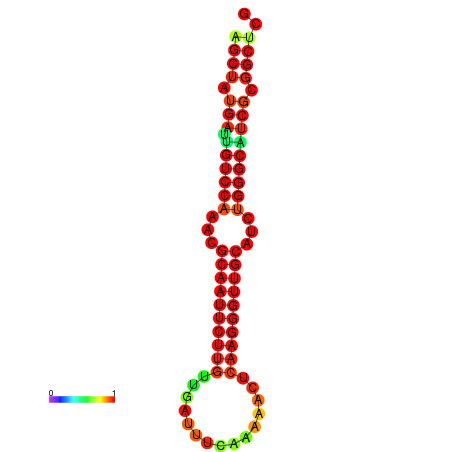 |
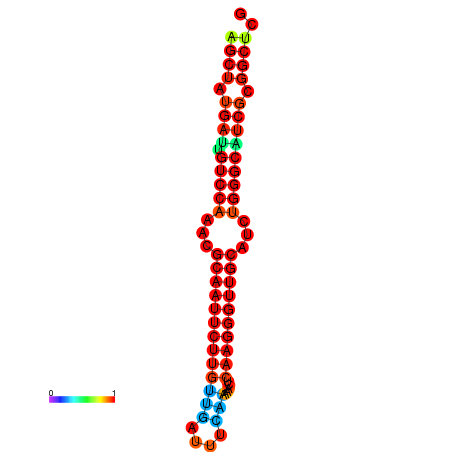 |
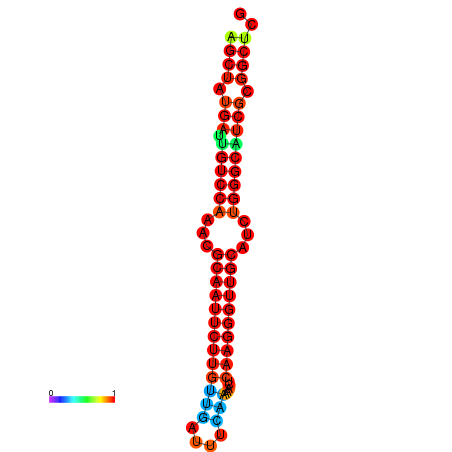 |
 |
Generated: 03/07/2013 at 04:53 PM