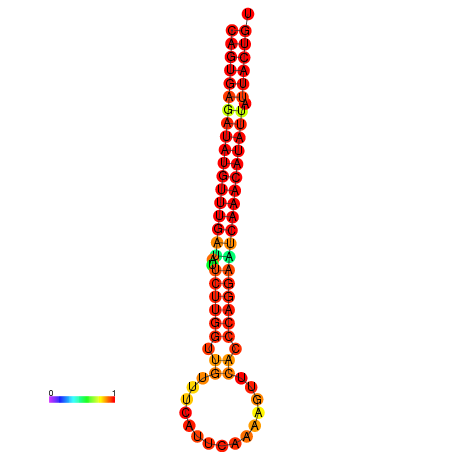
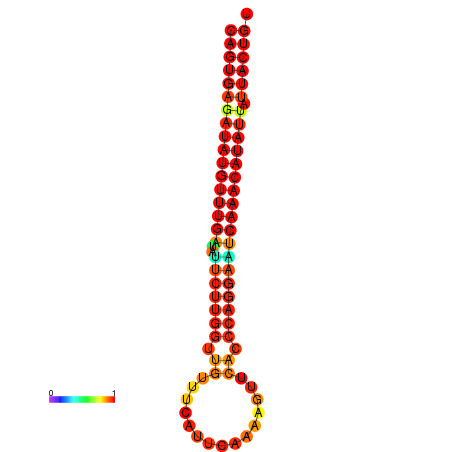
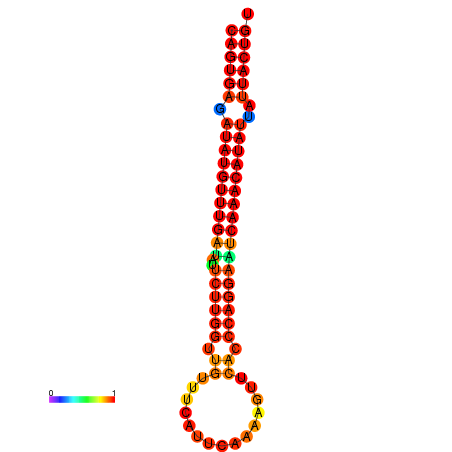
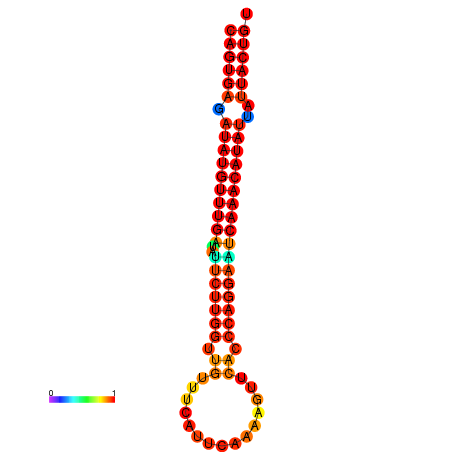
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:8564884-8565013 + | ACTT------------------------------------------GCCCTT-----------------------CAAAA---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droSim1 | chr3L:8005898-8006027 + | ACTT------------------------------------------GCCCTT-----------------------CAAAA---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTATCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droSec1 | super_0:844136-844265 + | ACTT------------------------------------------GCCCTT-----------------------CAAAA---------T-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droYak2 | chr3L:3348658-3348787 - | ACTA------------------------------------------GCCCTT-----------------------CAAAA---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droEre2 | scaffold_4784:5482466-5482595 - | ACTA------------------------------------------GCCCTT-----------------------CAAAT---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTAGCCTTCGCCAATCTACTAAAACA------- |
| droAna3 | scaffold_13337:8074872-8075002 - | ACTC------------------------------------------GCCTCT-----------------------CCAAA---------CACA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACTCTCGCCAATCTACTAAAATA------- |
| dp4 | chrXR_group3a:147689-147845 - | ACTA------------------------------------------GT---TAACCCTAACTCTCATCTGCCCACAAA--CCAACCCAAC-GA-------ACTAA-----------TTGGGGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAGAGTTCACCCAGGAATCAAACATATTATTACTGTGATACCCGGAAATCTACTAAACCC------- |
| droPer1 | super_23:151050-151206 - | ACTA------------------------------------------GT---TAACCCTAACTCTCATCTGCCCACAAA--CCAACCCAAC-GA-------ACTAA-----------TTGGGGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAGAGTTCACCCAGGAATCAAACATATTATTACTGTGATACCCGGAAATCTACTAAACCC------- |
| droWil1 | scaffold_180949:5214317-5214457 + | ACTT------------------------------------------GCCCT-----------CTCATCTGCCCACAAAAA---------C-GA-------ACTAA-----------TTGATGGTCCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATCGTCGG-AATCTACTAAAAAC------- |
| droVir3 | scaffold_13049:6215347-6215479 + | AC-------------------------------------------------------------------------CAAAT---------C-AA-------ACTACCCCCCACTCATCTAATGATTCCAGTAAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATTGTTGAAACTCTACTAAACCA------- |
| droMoj3 | scaffold_6680:4536880-4537026 + | AGCC------------------------------------------AC---------------------------CAAAATCAACCAAAC-TACCTATCCACTAC------CTCATCTAACGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATCGCTTAAACTCTACTAAAA-A------- |
| droGri2 | scaffold_15072:36258-36427 + | ACTACCAATAAAATAAAAATCCAACCAATCAACCAACCAACCAACTACCCA------------------------------------------------------------CTCATCTAATGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATTGTTGAAACTCTACTAAAAACAAATCAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:8564884-8565013 + | ACTT------------------------------------------GCCCTT-----------------------CAAAA---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droSim1 | chr3L:8005898-8006027 + | ACTT------------------------------------------GCCCTT-----------------------CAAAA---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTATCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droSec1 | super_0:844136-844265 + | ACTT------------------------------------------GCCCTT-----------------------CAAAA---------T-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droYak2 | chr3L:3348658-3348787 - | ACTA------------------------------------------GCCCTT-----------------------CAAAA---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACCCTCGCCAATCTACTAAAACA------- |
| droEre2 | scaffold_4784:5482466-5482595 - | ACTA------------------------------------------GCCCTT-----------------------CAAAT---------C-GA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTTCATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTAGCCTTCGCCAATCTACTAAAACA------- |
| droAna3 | scaffold_13337:8074872-8075002 - | ACTC------------------------------------------GCCTCT-----------------------CCAAA---------CACA-------ACTAA-----------TTGATGGTTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGACTCTCGCCAATCTACTAAAATA------- |
| dp4 | chrXR_group3a:147689-147845 - | ACTA------------------------------------------GT---TAACCCTAACTCTCATCTGCCCACAAA--CCAACCCAAC-GA-------ACTAA-----------TTGGGGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAGAGTTCACCCAGGAATCAAACATATTATTACTGTGATACCCGGAAATCTACTAAACCC------- |
| droPer1 | super_23:151050-151206 - | ACTA------------------------------------------GT---TAACCCTAACTCTCATCTGCCCACAAA--CCAACCCAAC-GA-------ACTAA-----------TTGGGGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAGAGTTCACCCAGGAATCAAACATATTATTACTGTGATACCCGGAAATCTACTAAACCC------- |
| droWil1 | scaffold_180949:5214317-5214457 + | ACTT------------------------------------------GCCCT-----------CTCATCTGCCCACAAAAA---------C-GA-------ACTAA-----------TTGATGGTCCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATCGTCGG-AATCTACTAAAAAC------- |
| droVir3 | scaffold_13049:6215347-6215479 + | AC-------------------------------------------------------------------------CAAAT---------C-AA-------ACTACCCCCCACTCATCTAATGATTCCAGTAAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATTGTTGAAACTCTACTAAACCA------- |
| droMoj3 | scaffold_6680:4536880-4537026 + | AGCC------------------------------------------AC---------------------------CAAAATCAACCAAAC-TACCTATCCACTAC------CTCATCTAACGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATCGCTTAAACTCTACTAAAA-A------- |
| droGri2 | scaffold_15072:36258-36427 + | ACTACCAATAAAATAAAAATCCAACCAATCAACCAACCAACCAACTACCCA------------------------------------------------------------CTCATCTAATGATTCCAGTGAGATATGTTTGATATTCTTGGTTGTTACATTCAAAAGTTCACCCAGGAATCAAACATATTATTACTGTGATTGTTGAAACTCTACTAAAAACAAATCAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
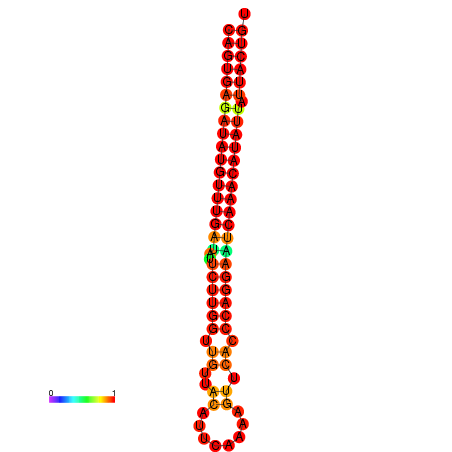
| dG=-25.5, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
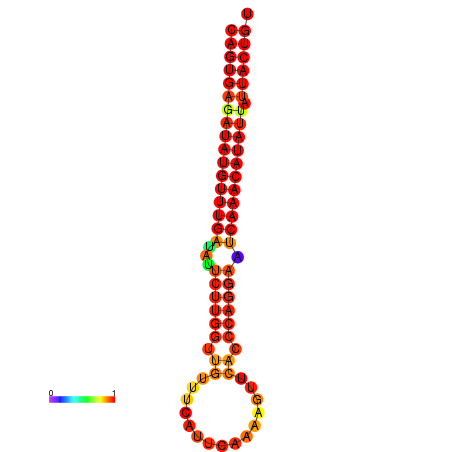
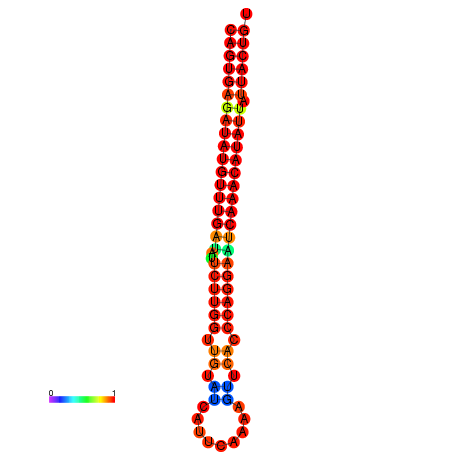
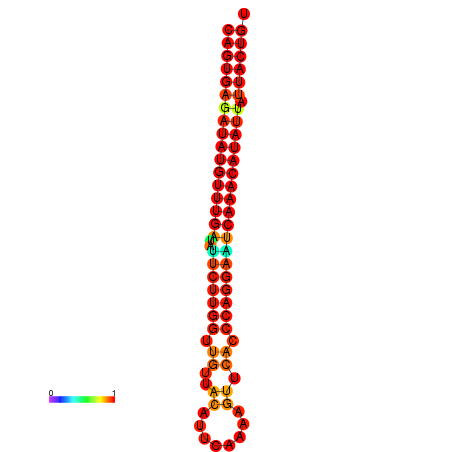
| dG=-25.5, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|---|
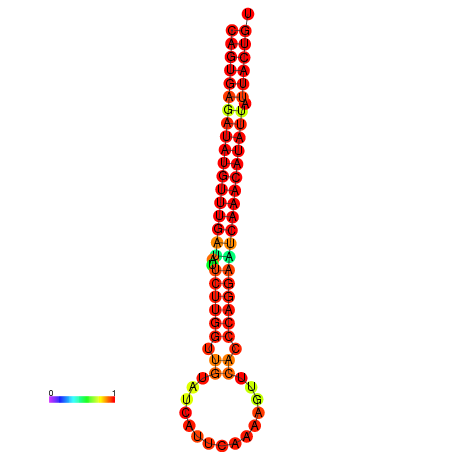 |
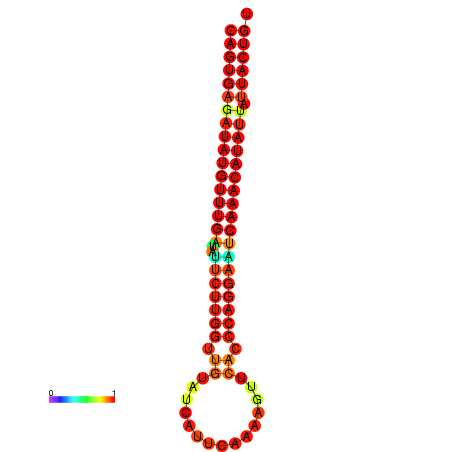 |
 |
 |
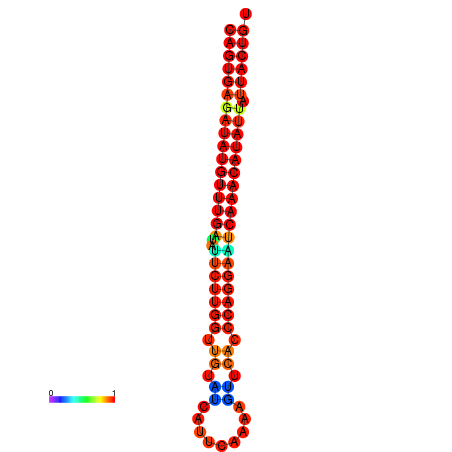 |
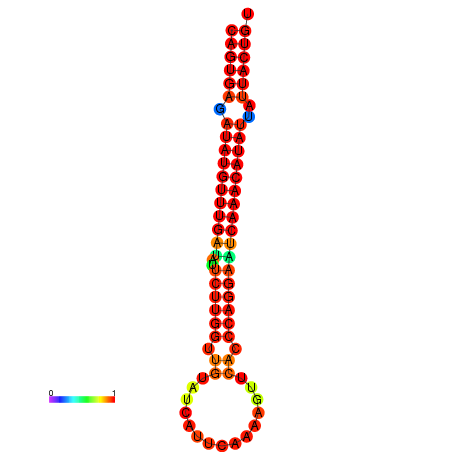
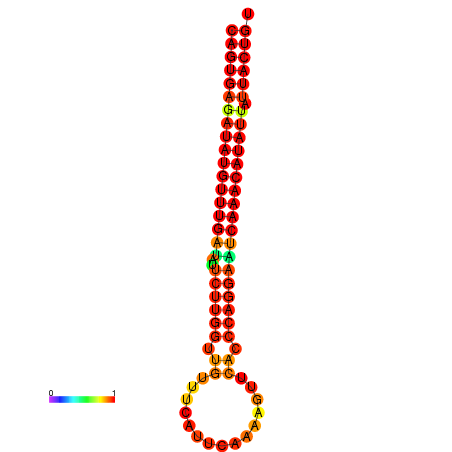
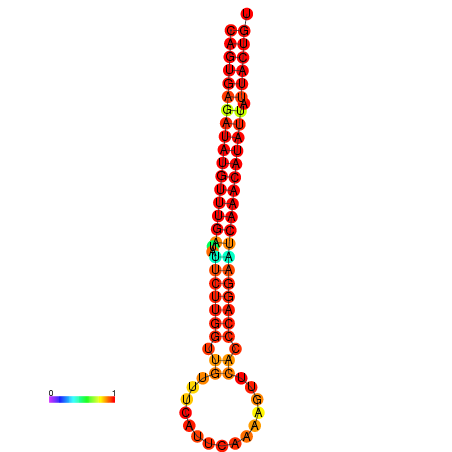
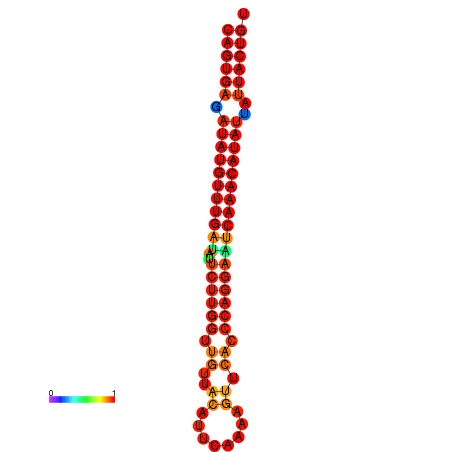
| dG=-25.5, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
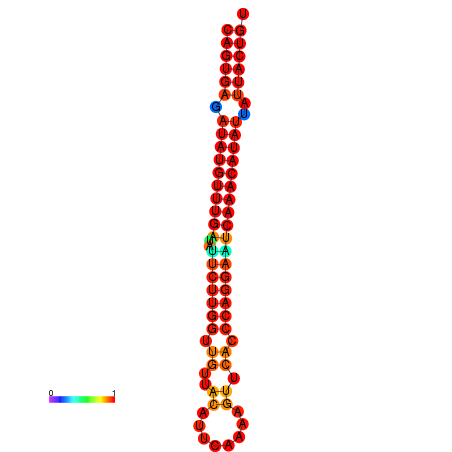
| dG=-25.5, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
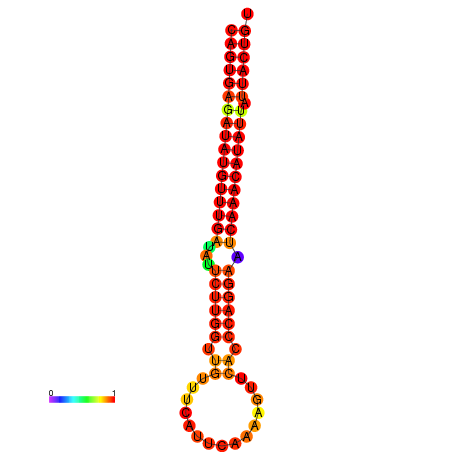 |
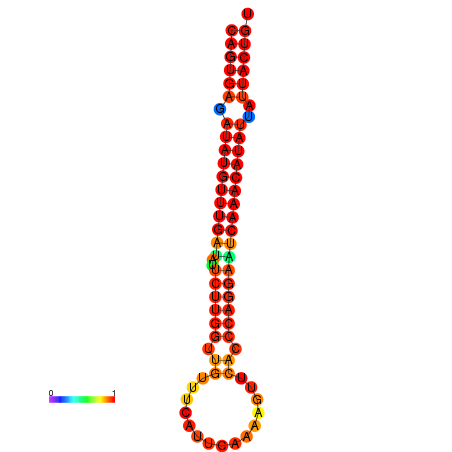
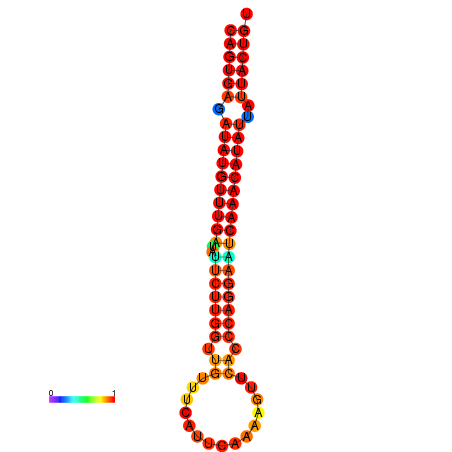
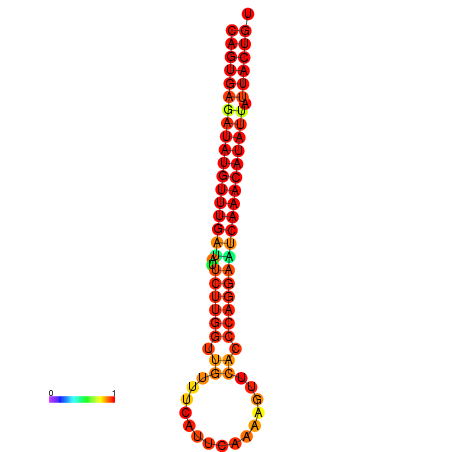
| dG=-25.5, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
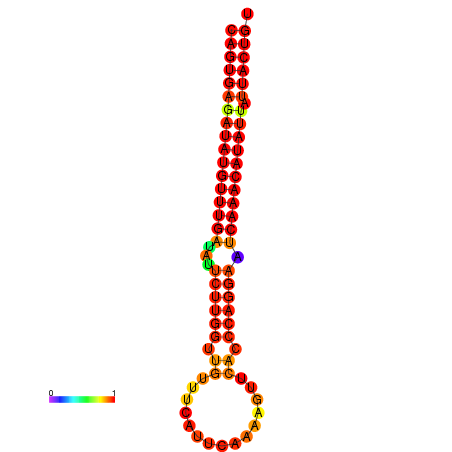 |
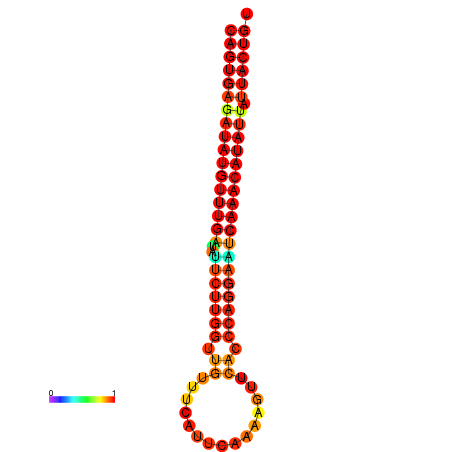
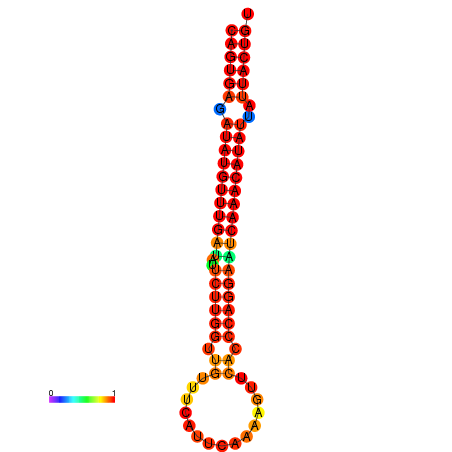
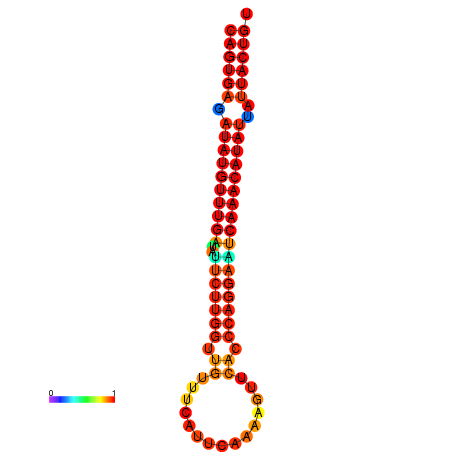
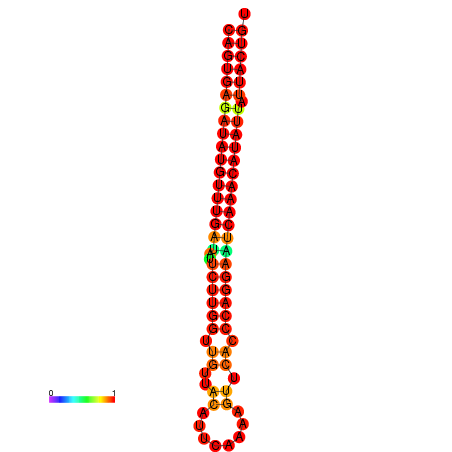
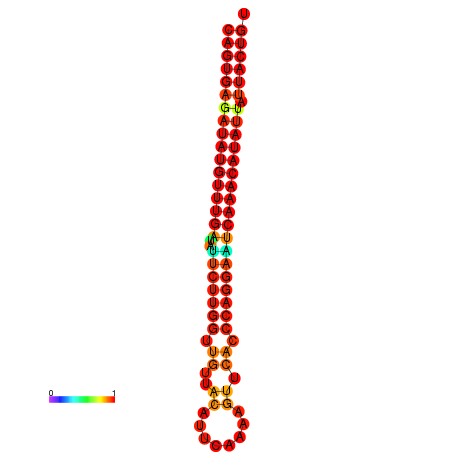
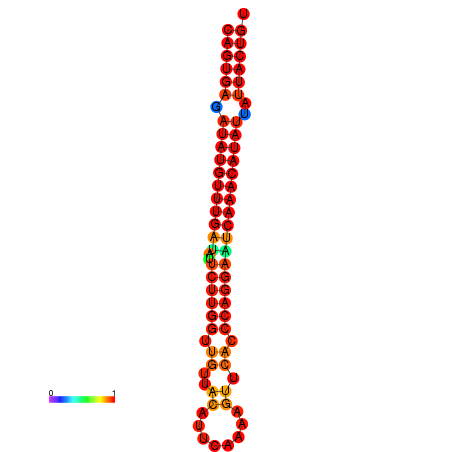
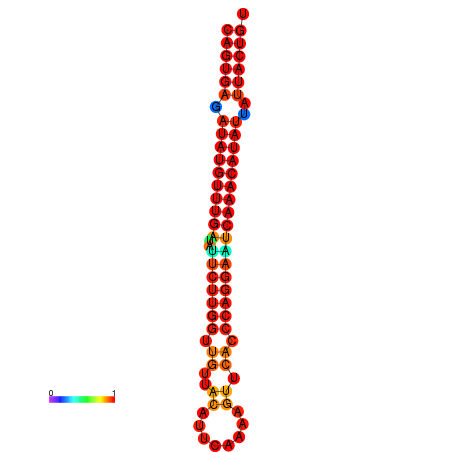
| dG=-26.8, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
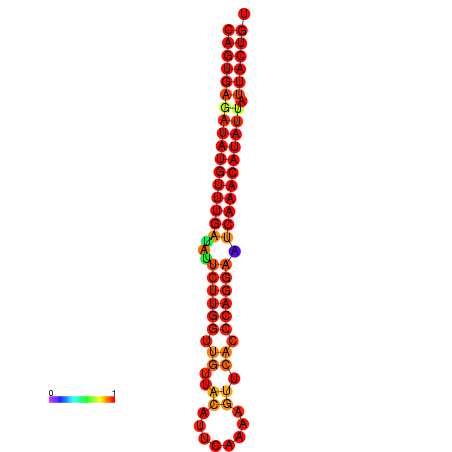 |
| dG=-26.8, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
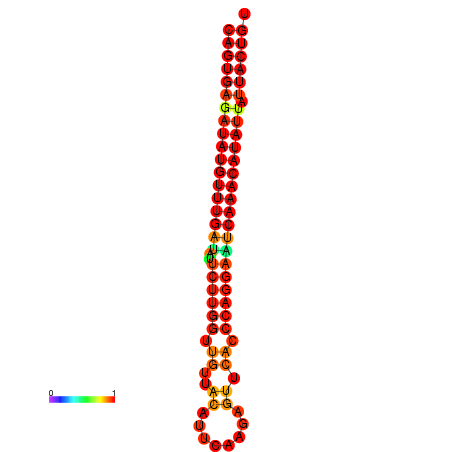 |
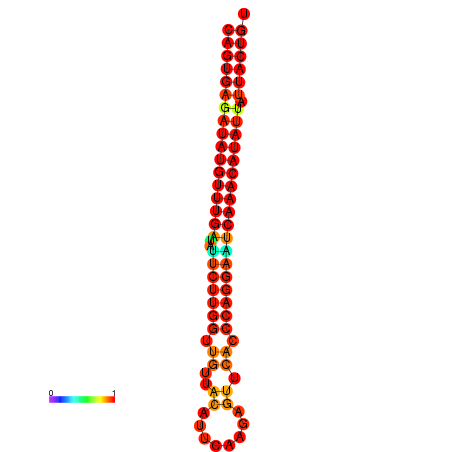 |
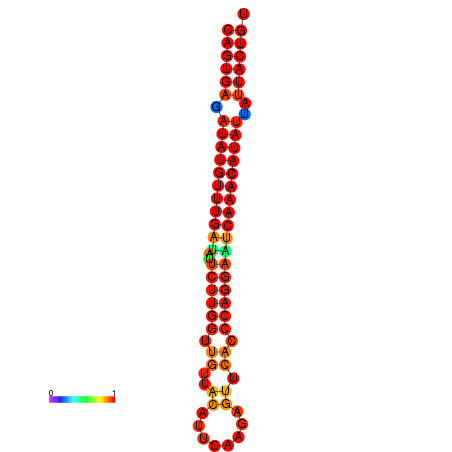 |
 |
 |
| dG=-26.8, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
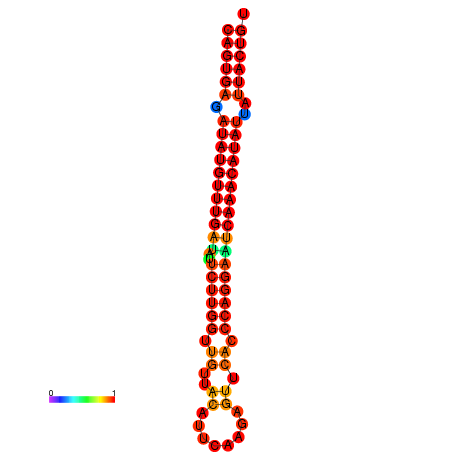 |
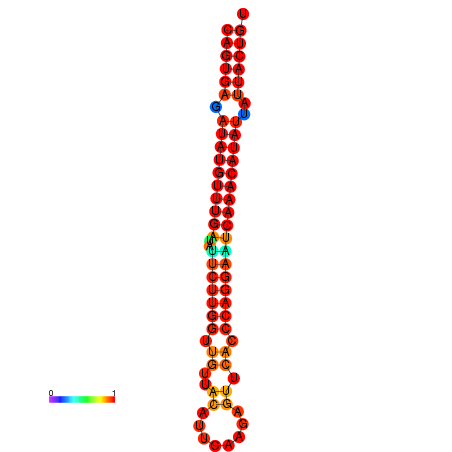 |
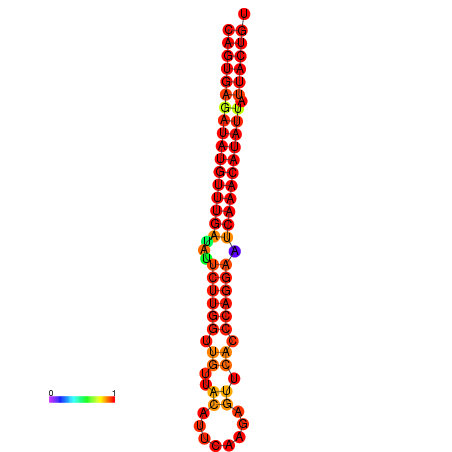 |
| dG=-26.8, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
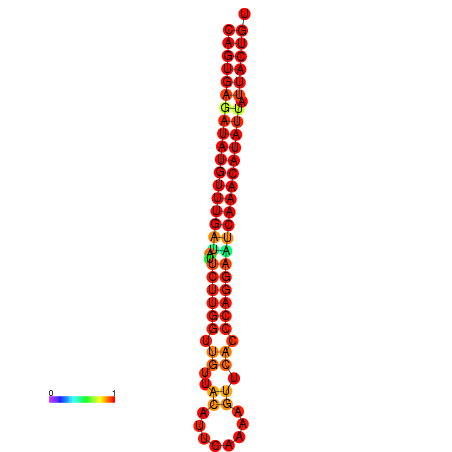 |
 |
 |
 |
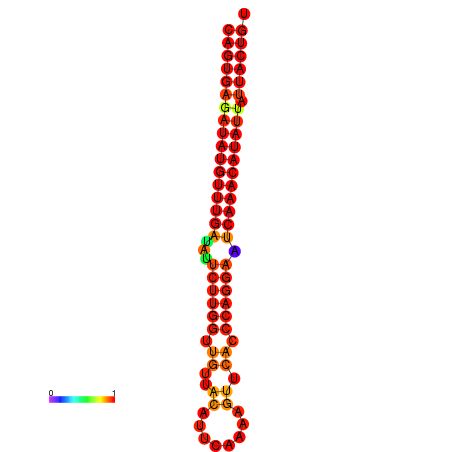 |
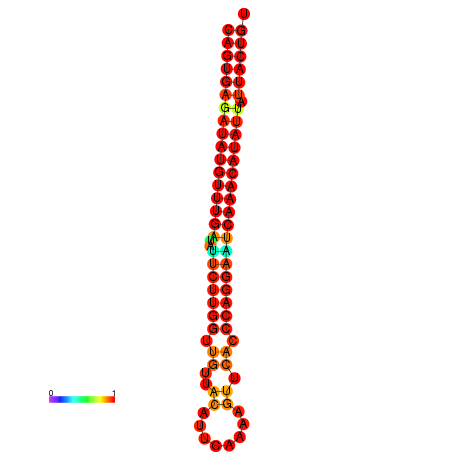
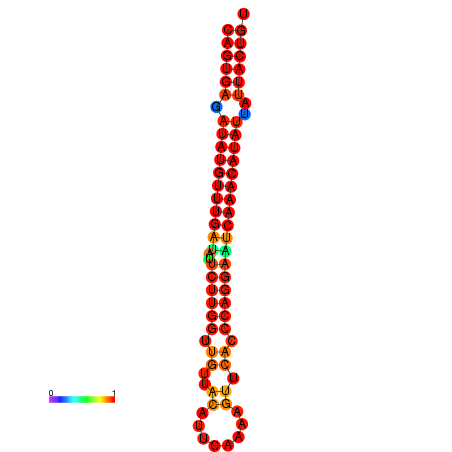
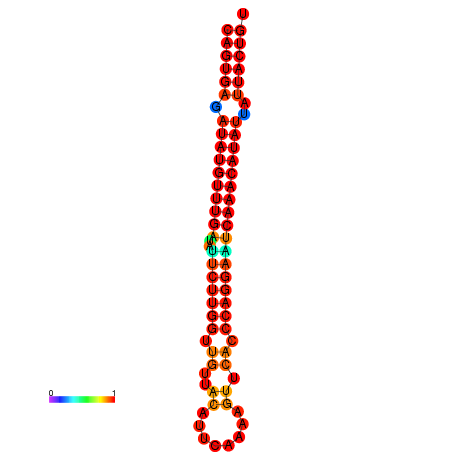
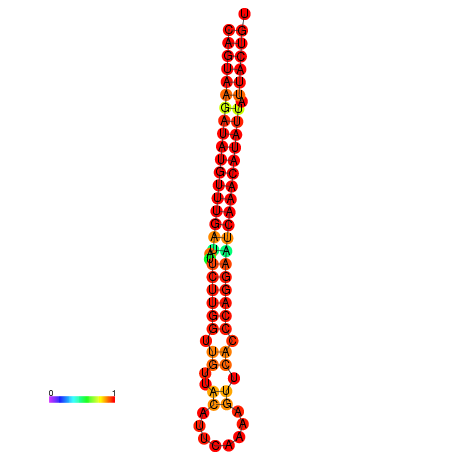
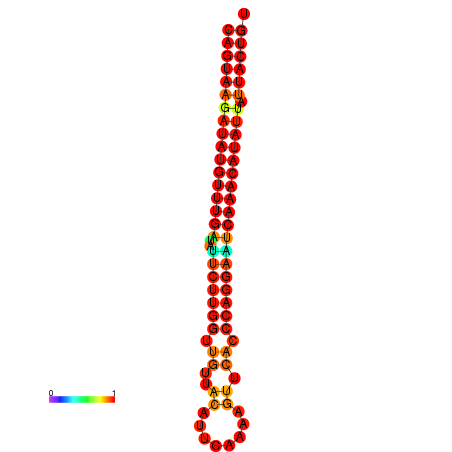
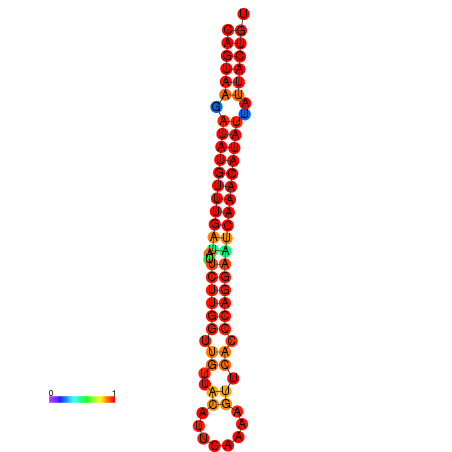
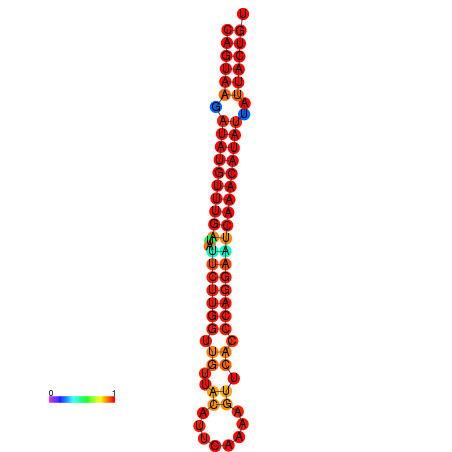
| dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.8, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.8, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:53 PM