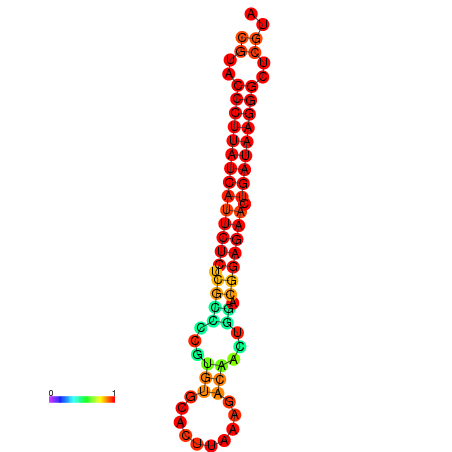
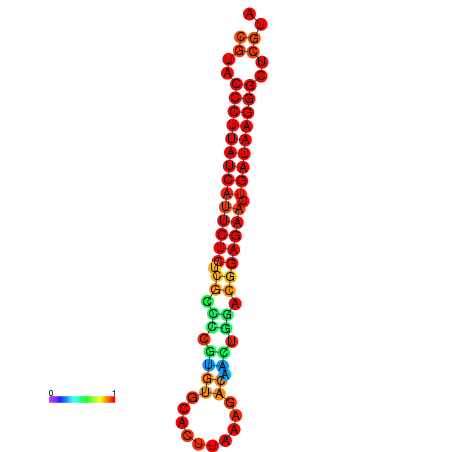
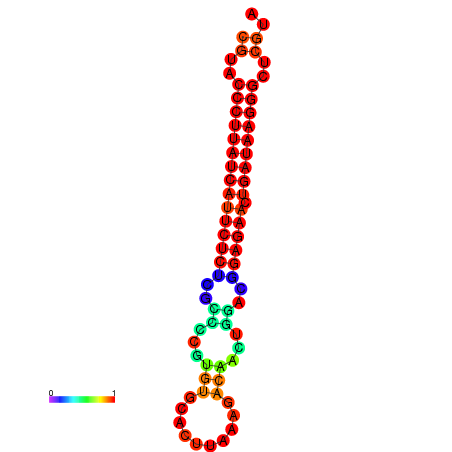
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:9216892-9217021 - | C--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCACTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAACCAATACCCA--------TT |
| droSim1 | chr2R:7673827-7673955 - | C--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCATTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAGC-AATCCCCA--------TT |
| droSec1 | super_1:6740004-6740132 - | T--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCATTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAGC-AATCTCCA--------TT |
| droYak2 | chr2R:7742247-7742374 + | C--TGGGCTG--------ACCCGAG-----------TGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCAT-TGAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAGC-AATCCCCA--------TT |
| droEre2 | scaffold_4845:16598085-16598213 + | C--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCATTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCGTCAAGC-AATCCCCA--------GT |
| droAna3 | scaffold_13266:16524568-16524693 - | CCCAGTGCCG--------G--CGAG------------TCCGGCCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATAATGAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAGGT-TG--TCCA--------TC |
| dp4 | chr3:696696-696816 + | ----GGGTCG--------GCCCGGGGACTCCG----GGTCGACCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATTAAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCATCA---------------------- |
| droPer1 | super_2:863138-863258 + | ----GGGTCG--------GCCCGGGGACTCCG----GGTCGACCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATTAAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCATCA---------------------- |
| droWil1 | scaffold_181009:2073162-2073280 - | A--CG----------------CGAAGAGTCAGTTAAAGCTGACCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATATCTAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCAACA---------------------- |
| droVir3 | scaffold_12875:243286-243412 + | T--TGGGCT-----------------------------TTGGCCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATATCTAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAAGTCAACTAAC-TAAGTGCAGCAACTATTT |
| droMoj3 | scaffold_6496:5255786-5255918 - | A--ATGGCCAATCTGCCGT--TGGG-----------CATTGGCCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATATCTATACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAAGTCAACTAA---ATACATG--------CG |
| droGri2 | scaffold_15245:10719551-10719668 - | C--CAGCCTG--------CATATGG-----------TTTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGGATAACTATACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCATCAAA-------------------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:9216892-9217021 - | C--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCACTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAACCAATACCCA--------TT |
| droSim1 | chr2R:7673827-7673955 - | C--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCATTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAGC-AATCCCCA--------TT |
| droSec1 | super_1:6740004-6740132 - | T--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCATTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAGC-AATCTCCA--------TT |
| droYak2 | chr2R:7742247-7742374 + | C--TGGGCTG--------ACCCGAG-----------TGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCAT-TGAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAAGC-AATCCCCA--------TT |
| droEre2 | scaffold_4845:16598085-16598213 + | C--TGGGCTG--------ACCCGAG-----------GGTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGCATTTAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCGTCAAGC-AATCCCCA--------GT |
| droAna3 | scaffold_13266:16524568-16524693 - | CCCAGTGCCG--------G--CGAG------------TCCGGCCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATAATGAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAATTCATCAGGT-TG--TCCA--------TC |
| dp4 | chr3:696696-696816 + | ----GGGTCG--------GCCCGGGGACTCCG----GGTCGACCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATTAAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCATCA---------------------- |
| droPer1 | super_2:863138-863258 + | ----GGGTCG--------GCCCGGGGACTCCG----GGTCGACCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATTAAAAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCATCA---------------------- |
| droWil1 | scaffold_181009:2073162-2073280 - | A--CG----------------CGAAGAGTCAGTTAAAGCTGACCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATATCTAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCAACA---------------------- |
| droVir3 | scaffold_12875:243286-243412 + | T--TGGGCT-----------------------------TTGGCCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATATCTAGACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAAGTCAACTAAC-TAAGTGCAGCAACTATTT |
| droMoj3 | scaffold_6496:5255786-5255918 - | A--ATGGCCAATCTGCCGT--TGGG-----------CATTGGCCGGTGCATTCGTACCCTTATCATTCTCGCGCCCCGTGTGCATATCTATACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAAGTCAACTAA---ATACATG--------CG |
| droGri2 | scaffold_15245:10719551-10719668 - | C--CAGCCTG--------CATATGG-----------TTTTGGCCGGTGCATTCGTACCCTTATCATTCTCTCGCCCCGTGTGGATAACTATACAACTGGACGGAGAACTGATAAGGGCTCGTATCACCAACTCATCAAA-------------------- |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|
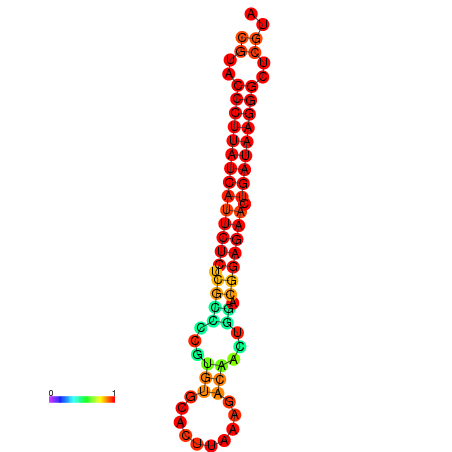 |
 |
 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|
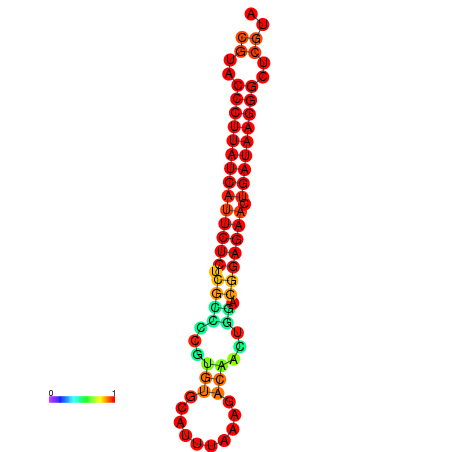 |
 |
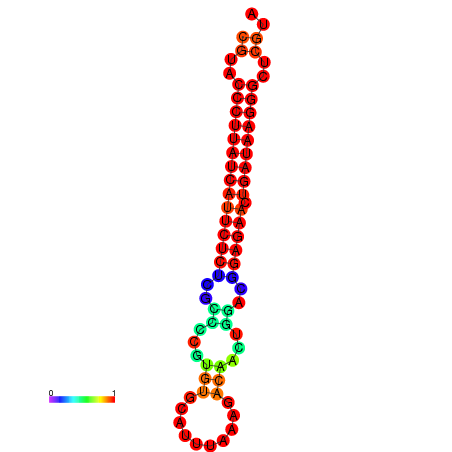 |
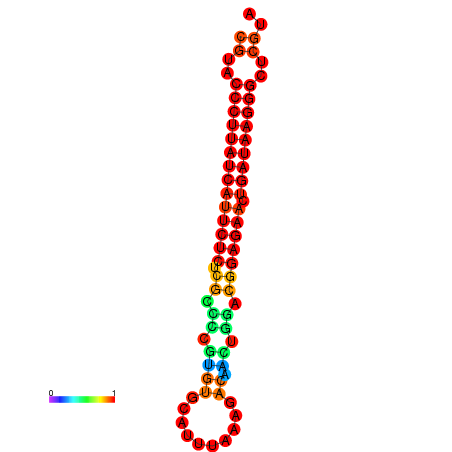
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|
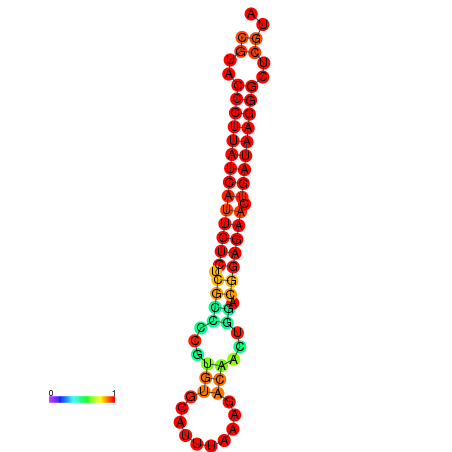 |
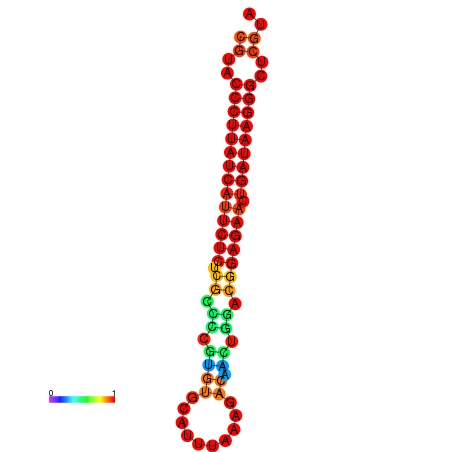 |
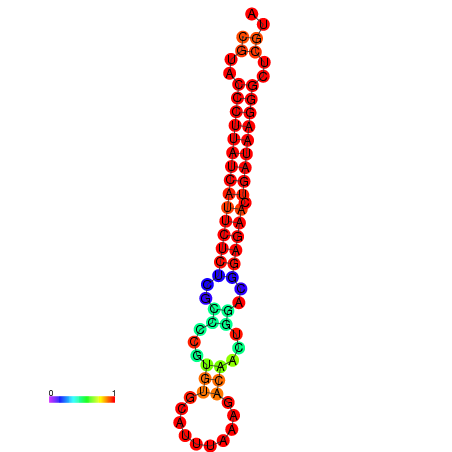 |
| dG=-27.3, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-26.6, p-value=0.009901 |
|---|---|---|
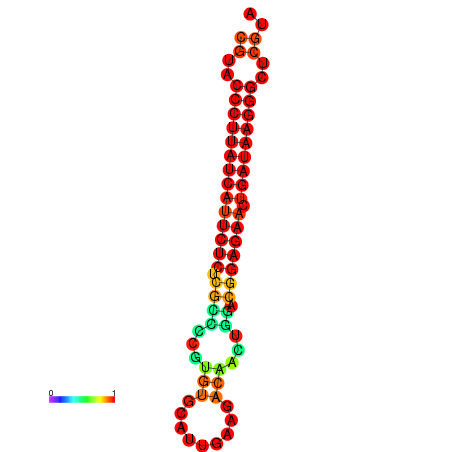 |
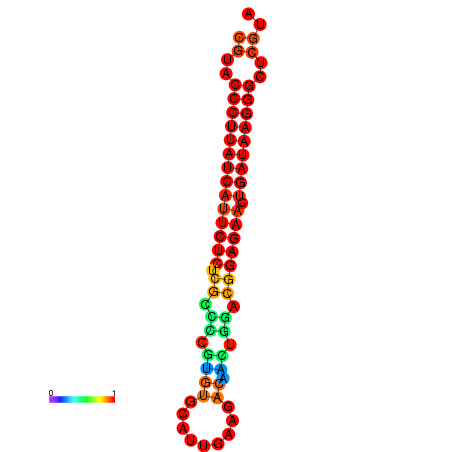 |
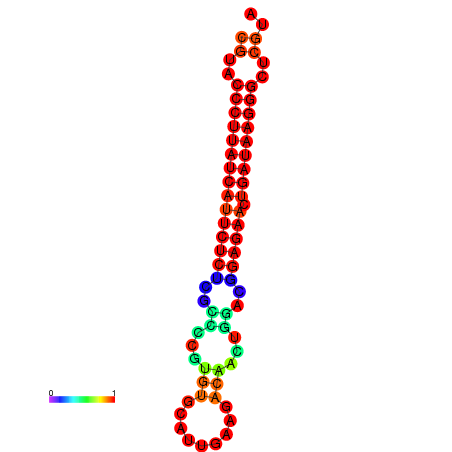 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|
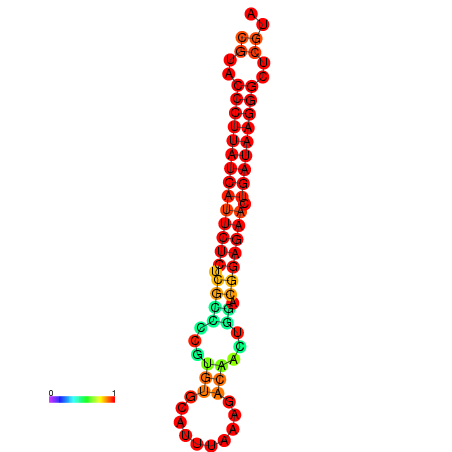 |
 |
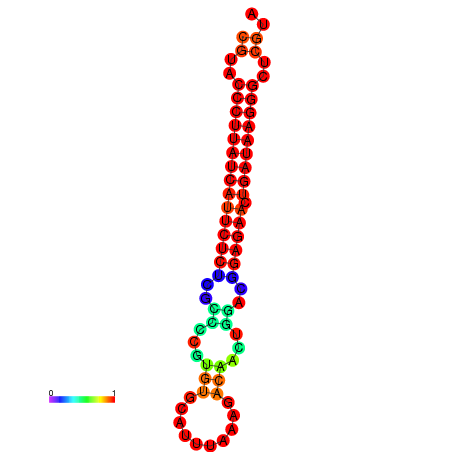 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.2, p-value=0.009901 |
|---|---|---|---|
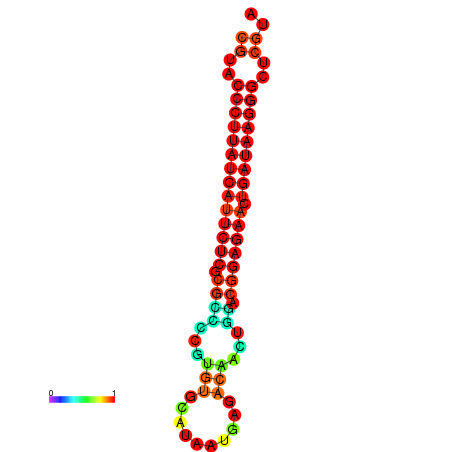 |
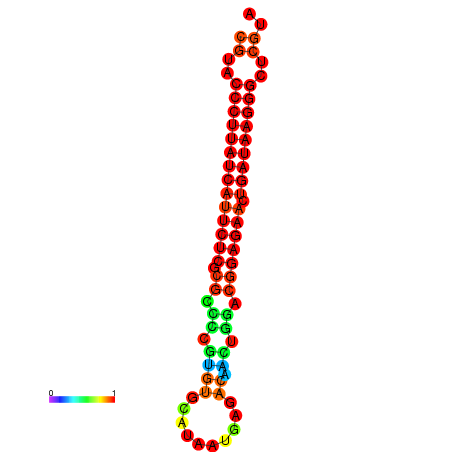 |
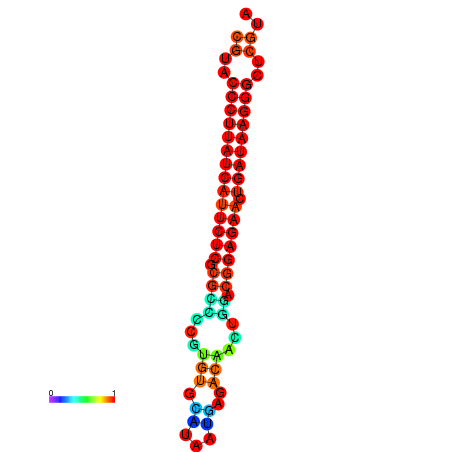 |
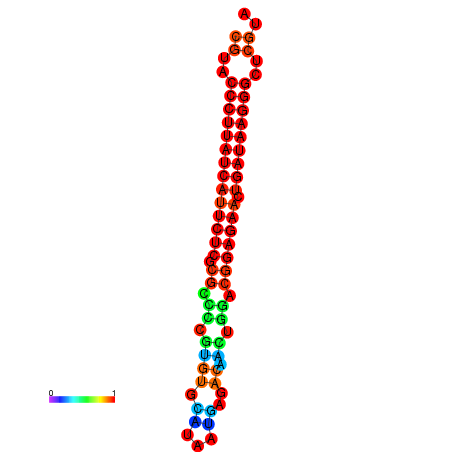 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 |
|---|---|
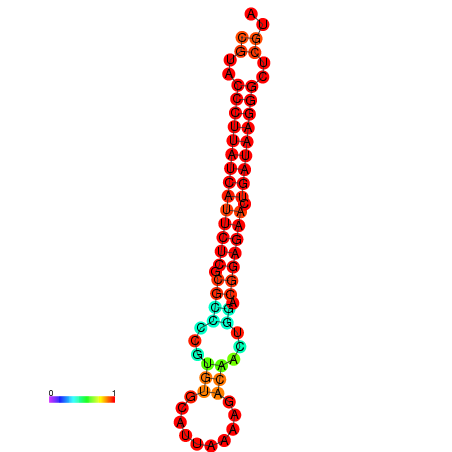 |
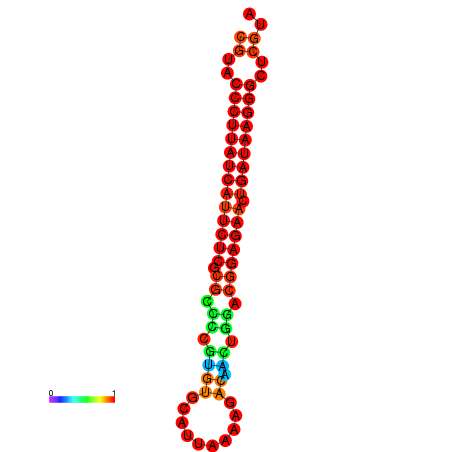 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 |
|---|---|
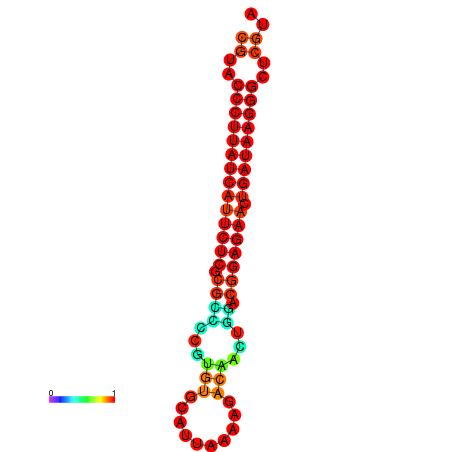 |
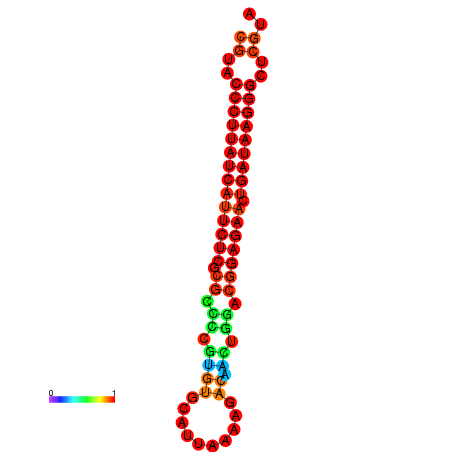 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|---|
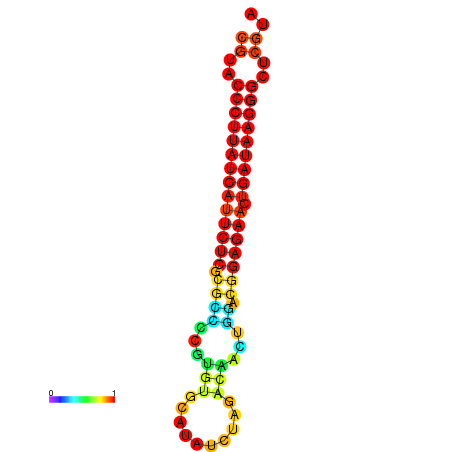 |
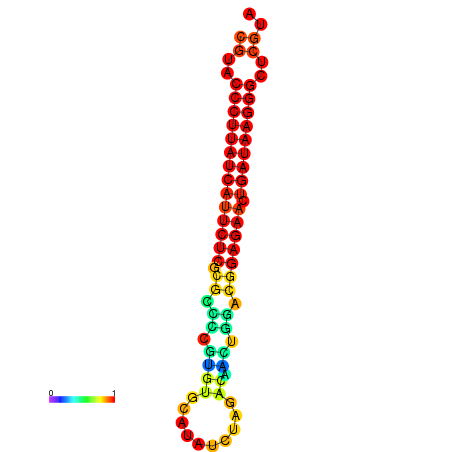 |
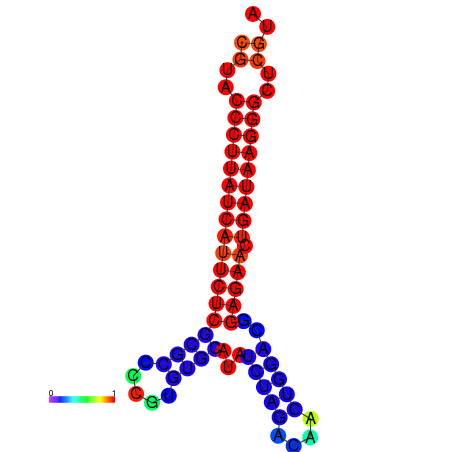 |
| dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|---|
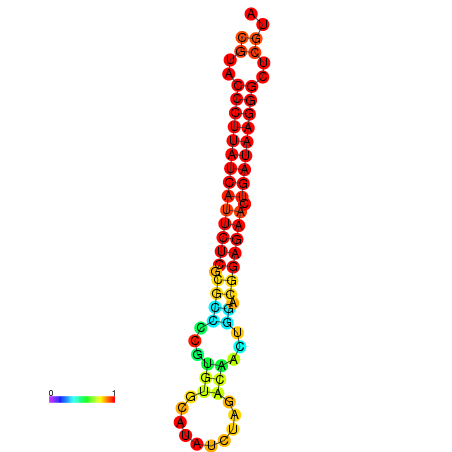 |
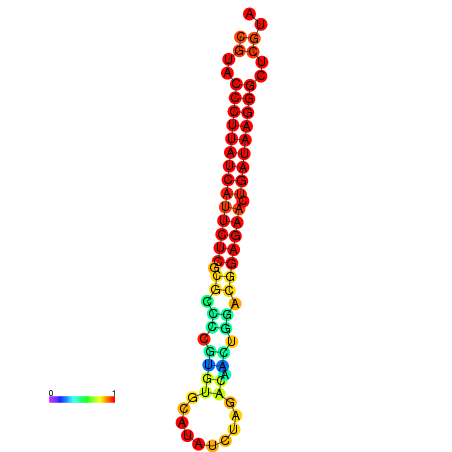 |
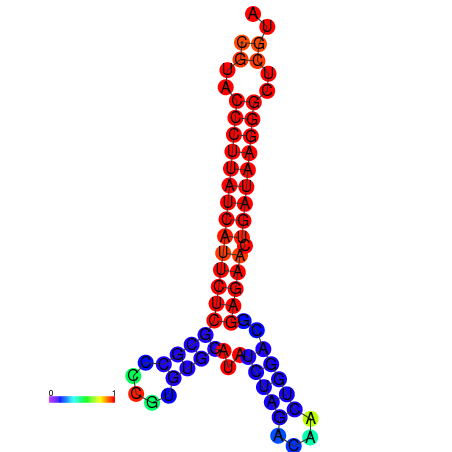 |
| dG=-27.8, p-value=0.009901 | dG=-27.5, p-value=0.009901 | dG=-27.0, p-value=0.009901 |
|---|---|---|
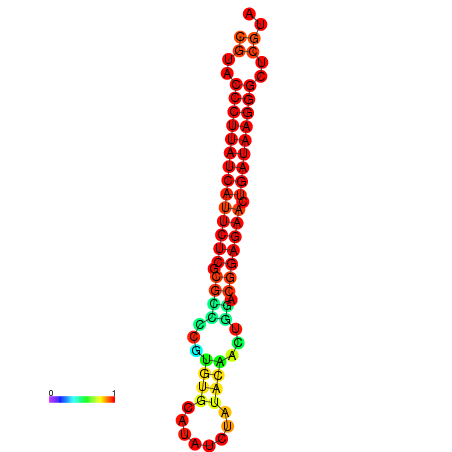 |
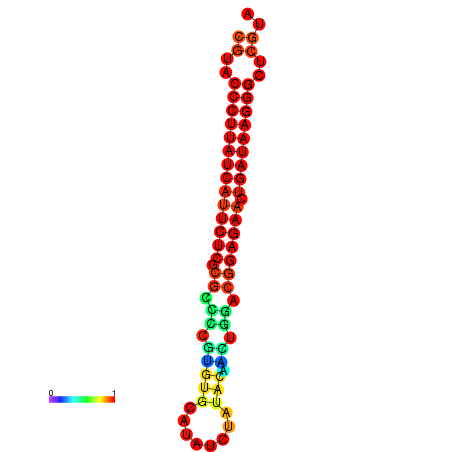 |
 |
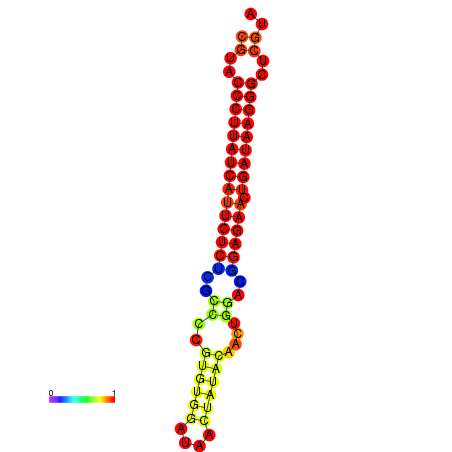
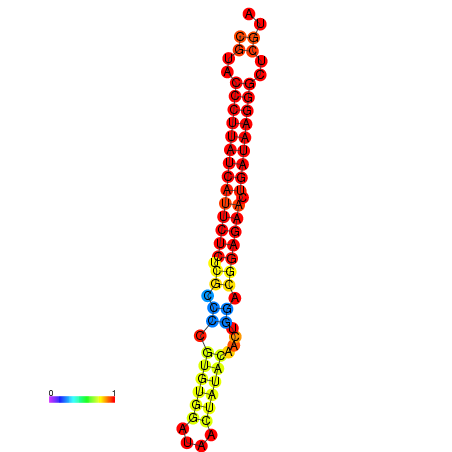
| dG=-29.8, p-value=0.009901 | dG=-29.1, p-value=0.009901 | dG=-29.1, p-value=0.009901 |
|---|---|---|
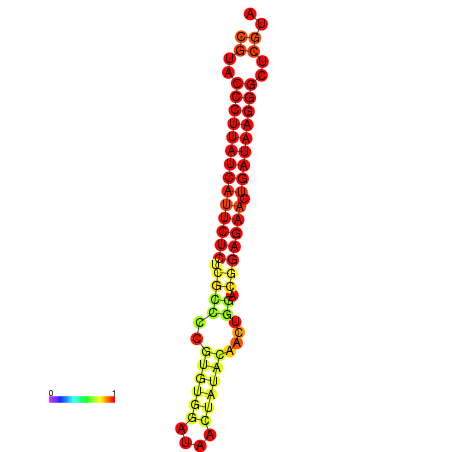 |
 |
 |
Generated: 03/07/2013 at 04:52 PM