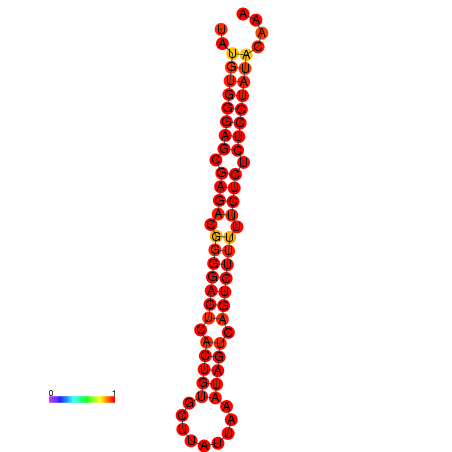
| ACGTACTGCATTC-ATGTGGGAGCGAGACGGGGACTCACTGTGCTTTTGAAGTAGTCAGTCTTTTTCTCTCTCCTATACAAATTGCGGGCGTA | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| .......................................................TCAGTCTTTTTCTCTCTCCTAT................ | 22 | 2 | 17802.50 | 35605 | 18647 | 16958 |
| .......................................................TCAGTCTTTTTCTCTCTCCTA................. | 21 | 2 | 2349.50 | 4699 | 0 | 4699 |
| .......................................................TCAGTCTTTTTCTCTCTCCT.................. | 20 | 2 | 2287.50 | 4575 | 0 | 4575 |
| .........................................................AGTCTTTTTCTCTCTCCTAT................ | 20 | 2 | 267.00 | 534 | 304 | 230 |
| .......................................................TCAGTCTTTTTCTCTCTCCTATA............... | 23 | 2 | 186.50 | 373 | 209 | 164 |
| .......................................................TCAGTCTTTTTCTCTCTCC................... | 19 | 2 | 44.50 | 89 | 0 | 89 |
| .........................................................AGTCTTTTTCTCTCTCCT.................. | 18 | 2 | 26.50 | 53 | 0 | 53 |
| ..................GGGAGCGAGACGGGGACTCACT..................................................... | 22 | 2 | 24.50 | 49 | 0 | 49 |
| ..................GGGAGCGAGACGGGGACTCAC...................................................... | 21 | 2 | 22.00 | 44 | 34 | 10 |
| ........................................................CAGTCTTTTTCTCTCTCCTA................. | 20 | 2 | 8.50 | 17 | 17 | 0 |
| ........................................................CAGTCTTTTTCTCTCTCCT.................. | 19 | 2 | 7.00 | 14 | 2 | 12 |
| ..................GGGAGCGAGACGGGGACT......................................................... | 18 | 2 | 7.00 | 14 | 0 | 14 |
| ........................................................CAGTCTTTTTCTCTCTCCTATA............... | 22 | 2 | 5.00 | 10 | 7 | 3 |
| .......................................................TCAGTCTTTTTCTCTCTC.................... | 18 | 2 | 4.00 | 8 | 8 | 0 |
| ..................GGGAGCGAGACGGGGACTCA....................................................... | 20 | 2 | 3.50 | 7 | 6 | 1 |
| ...................GGAGCGAGACGGGGACTCACT..................................................... | 21 | 2 | 3.00 | 6 | 0 | 6 |
| .................TGGGAGCGAGACGGGGAC.......................................................... | 18 | 2 | 2.50 | 5 | 0 | 5 |
| .........................................................AGTCTTTTTCTCTCTCCTATA............... | 21 | 2 | 2.50 | 5 | 5 | 0 |
| .................TGGGAGCGAGACGGGGACTCAC...................................................... | 22 | 2 | 1.50 | 3 | 1 | 2 |
| .......................................................TCAGTCTTTTTCTCTCTCCTATAC.............. | 24 | 2 | 1.00 | 2 | 2 | 0 |