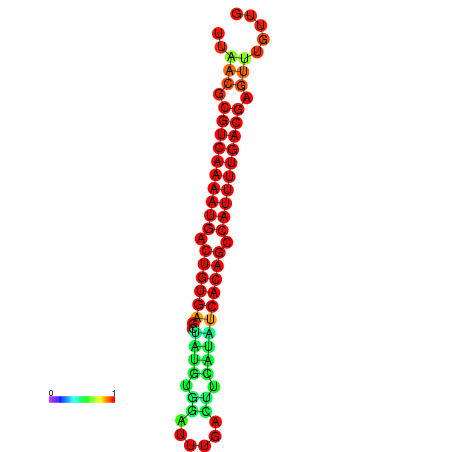

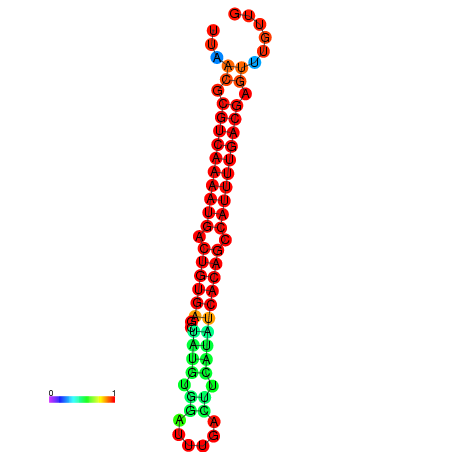
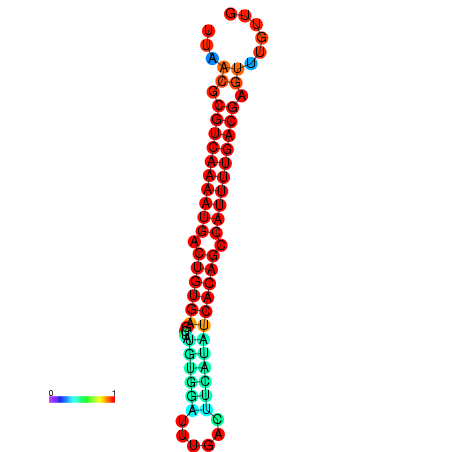

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:8985248-8985347 + | TCTA---CCTCT------------------GCT------------------GTCTATTAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GACAG-CCAGCCT |
| droSim1 | chrX_random:2632466-2632565 + | TCTG---CCTCT------------------GCT------------------GTCTATCAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GGCAG-CCAGCCT |
| droSec1 | super_34:345603-345702 + | CCTG---CCTCT------------------GCT------------------GTCTATCAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GGCAG-CCAGCCT |
| droYak2 | chrX:12330556-12330651 - | CTTA---CCCCT------------------GCT------------------GTCTAGTAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGCTG-GCCAGT-----CT |
| droEre2 | scaffold_4690:6132295-6132393 - | TTTAAACCCCCT------------------GCT------------------GTCTATTAGCGCGTCAAAGTGACTGTGAACTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GGCAGC-C----C |
| droAna3 | scaffold_13047:797460-797571 + | TGTCCCGCTTCT------------------GCTT----CATTAC--CT---GTTTCCCAGCGCGTCGAAATGACTGTGACTTATGCGT----ATTTGA-TGTCATATCACAGCCATTTTGACGAGTTTGGTG-GACAGT-GAAAGT |
| dp4 | chrXL_group1e:7000785-7000885 - | GCTA---CTGCT------------------GCT------------------GCACATTTACGCGTCAAAATGACTGTGAGCTATGTGT----TTTTGA-TCTCATATCACAGCCATTTTGACGAGTTTGATG-TGCAGTCCCACTC |
| droPer1 | super_13:1553253-1553353 + | GCTA---CTGCT------------------GCT------------------GCACATTTACGCGTCAAAATGACTGTGAGCTATGTGT----TTTTGA-TCTCATATCACAGCCATTTTGACGAGTTTGATG-TGCAGTCCCACTC |
| droWil1 | scaffold_180777:2053037-2053171 + | TCCC---CCAATGTACCTACCCCTGCAACTGCTGTGCTT---ACAACTGATGTCCATGGTCGCGTCAAAATGACTGTGAGCTATGTTAACCGGTCAGAAATTCATATCACAGCCATTTTGACGAGTTCGATG-GGCAT-AC---GA |
| droVir3 | scaffold_12970:9304312-9304422 + | TCTC---TCGCT------------------GCT---------GCCAATGCTCCATATTGGCGCGTCAAAATGACTGTGAGCTATGTTGTTGCATTTGCCATTCATATCACAGCCATTTTGACGAGTTTGATGTGGCAGC-CA---- |
| droMoj3 | scaffold_6473:13342549-13342650 - | TCCT---CTAAT------------------GCT------------------CCATATTTGCGCGTCAAATTGACTGTGAGCTATGTTT----ATGAGATTATCATATCACAGCCATTTTGACGAGTTTGATGTGGCAG-CCACCAA |
| droGri2 | scaffold_15203:306998-307104 + | TCG-------TT------------------GCT---------ATCAGTGCTCCATATTGGCGCGTCAAATTGACTGTGAGCTATGTTGTTGCAGTTGTAATTCATATCACAGCCATTTTGACGAGTTTGATGTGGCAGC-CA---- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:8985248-8985347 + | TCTA---CCTCT------------------GCT------------------GTCTATTAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GACAG-CCAGCCT |
| droSim1 | chrX_random:2632466-2632565 + | TCTG---CCTCT------------------GCT------------------GTCTATCAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GGCAG-CCAGCCT |
| droSec1 | super_34:345603-345702 + | CCTG---CCTCT------------------GCT------------------GTCTATCAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GGCAG-CCAGCCT |
| droYak2 | chrX:12330556-12330651 - | CTTA---CCCCT------------------GCT------------------GTCTAGTAACGCGTCAAAATGACTGTGAGCTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGCTG-GCCAGT-----CT |
| droEre2 | scaffold_4690:6132295-6132393 - | TTTAAACCCCCT------------------GCT------------------GTCTATTAGCGCGTCAAAGTGACTGTGAACTATGTGG----ATTTGA-CTTCATATCACAGCCATTTTGACGAGTTTGTTG-GGCAGC-C----C |
| droAna3 | scaffold_13047:797460-797571 + | TGTCCCGCTTCT------------------GCTT----CATTAC--CT---GTTTCCCAGCGCGTCGAAATGACTGTGACTTATGCGT----ATTTGA-TGTCATATCACAGCCATTTTGACGAGTTTGGTG-GACAGT-GAAAGT |
| dp4 | chrXL_group1e:7000785-7000885 - | GCTA---CTGCT------------------GCT------------------GCACATTTACGCGTCAAAATGACTGTGAGCTATGTGT----TTTTGA-TCTCATATCACAGCCATTTTGACGAGTTTGATG-TGCAGTCCCACTC |
| droPer1 | super_13:1553253-1553353 + | GCTA---CTGCT------------------GCT------------------GCACATTTACGCGTCAAAATGACTGTGAGCTATGTGT----TTTTGA-TCTCATATCACAGCCATTTTGACGAGTTTGATG-TGCAGTCCCACTC |
| droWil1 | scaffold_180777:2053037-2053171 + | TCCC---CCAATGTACCTACCCCTGCAACTGCTGTGCTT---ACAACTGATGTCCATGGTCGCGTCAAAATGACTGTGAGCTATGTTAACCGGTCAGAAATTCATATCACAGCCATTTTGACGAGTTCGATG-GGCAT-AC---GA |
| droVir3 | scaffold_12970:9304312-9304422 + | TCTC---TCGCT------------------GCT---------GCCAATGCTCCATATTGGCGCGTCAAAATGACTGTGAGCTATGTTGTTGCATTTGCCATTCATATCACAGCCATTTTGACGAGTTTGATGTGGCAGC-CA---- |
| droMoj3 | scaffold_6473:13342549-13342650 - | TCCT---CTAAT------------------GCT------------------CCATATTTGCGCGTCAAATTGACTGTGAGCTATGTTT----ATGAGATTATCATATCACAGCCATTTTGACGAGTTTGATGTGGCAG-CCACCAA |
| droGri2 | scaffold_15203:306998-307104 + | TCG-------TT------------------GCT---------ATCAGTGCTCCATATTGGCGCGTCAAATTGACTGTGAGCTATGTTGTTGCAGTTGTAATTCATATCACAGCCATTTTGACGAGTTTGATGTGGCAGC-CA---- |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.6, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
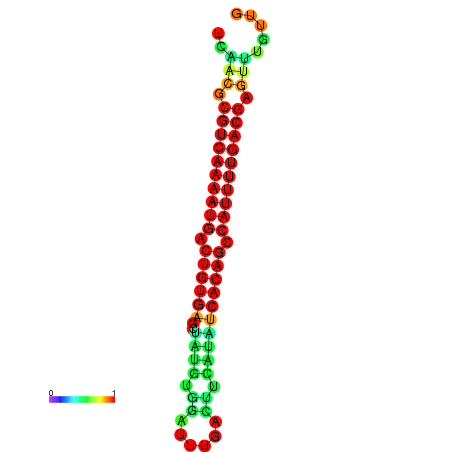 |
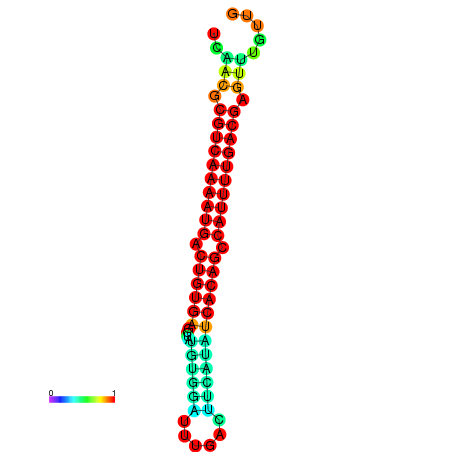 |
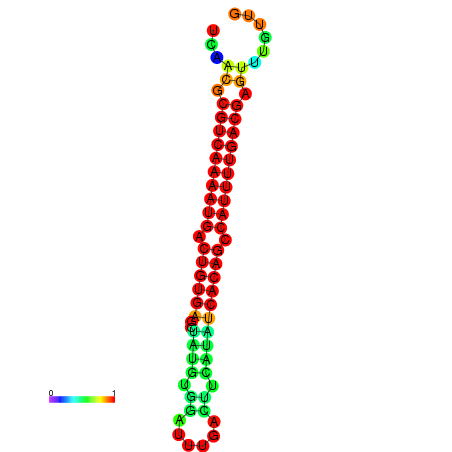 |
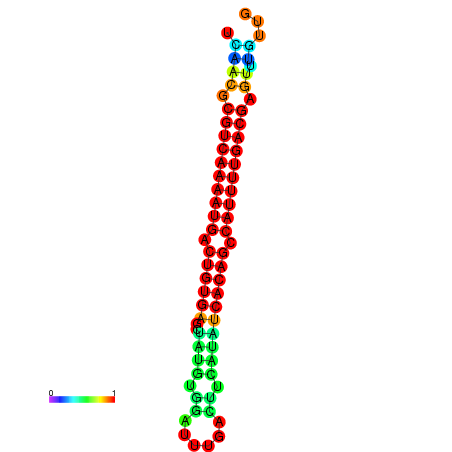 |
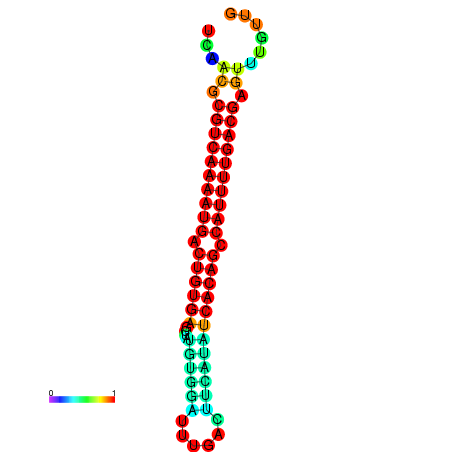 |
| dG=-26.6, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
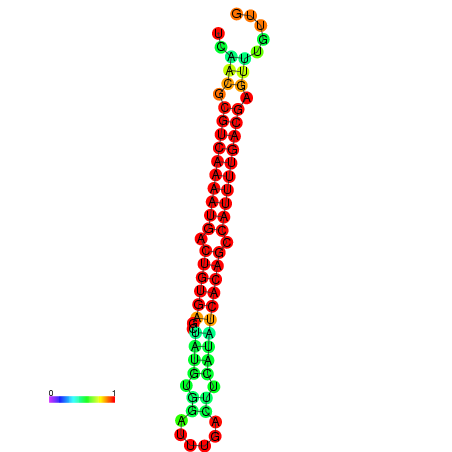 |
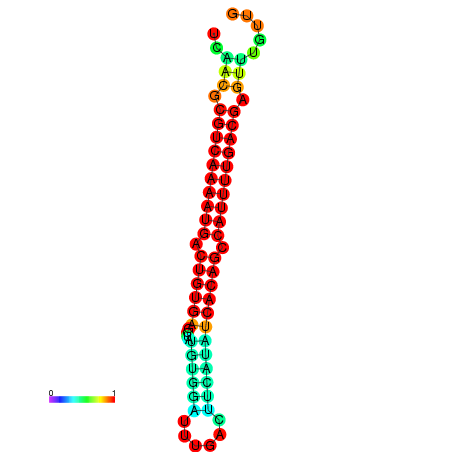 |
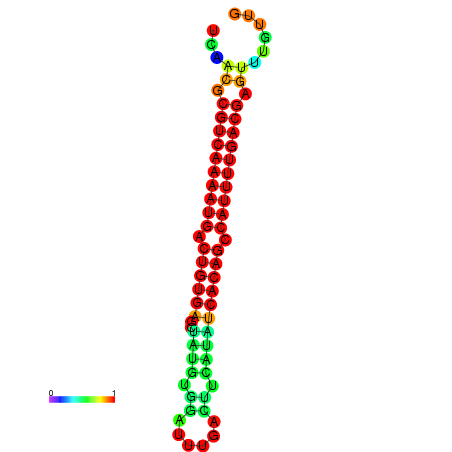 |
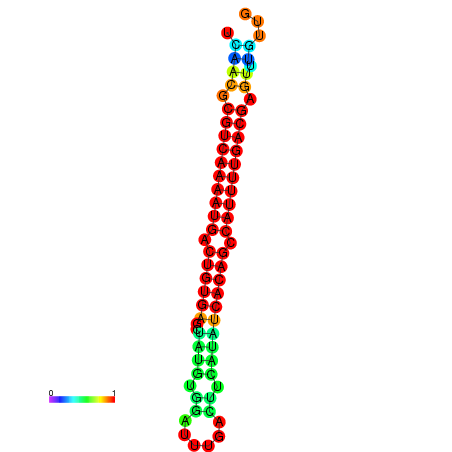 |
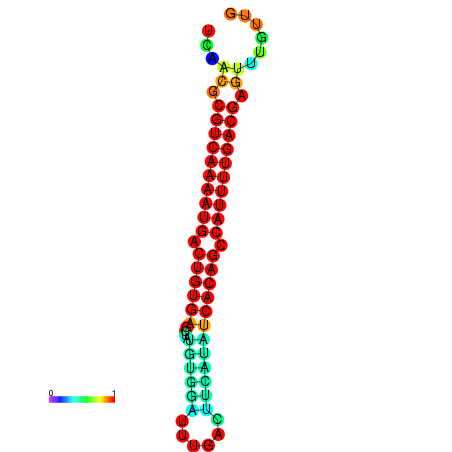 |
| dG=-26.7, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|---|---|
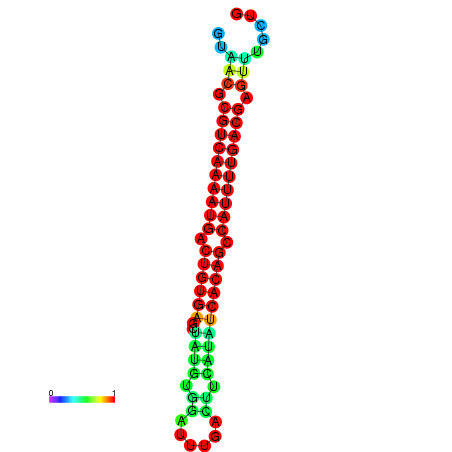 |
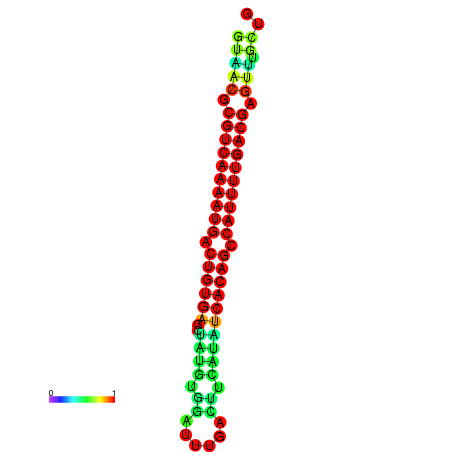 |
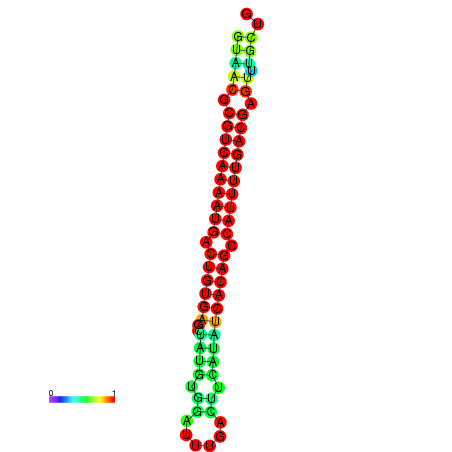 |
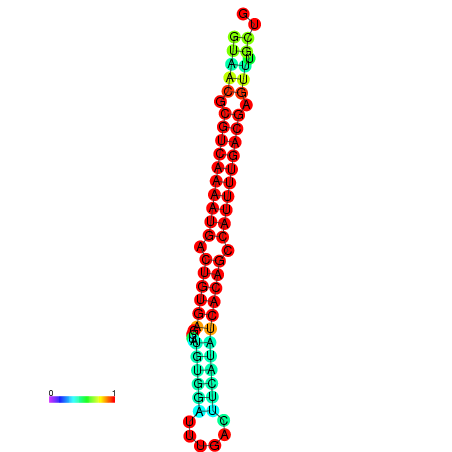 |
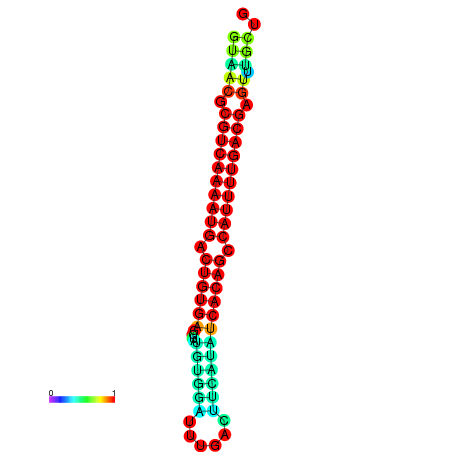 |
| dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|---|
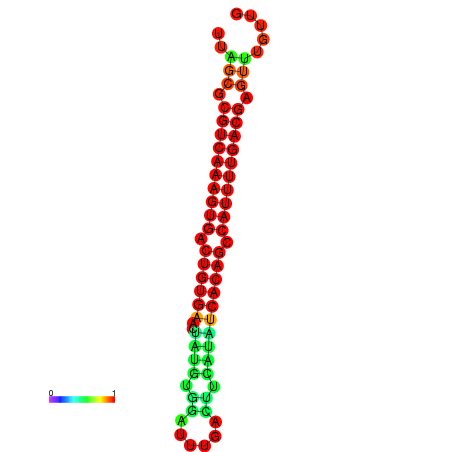 |
 |
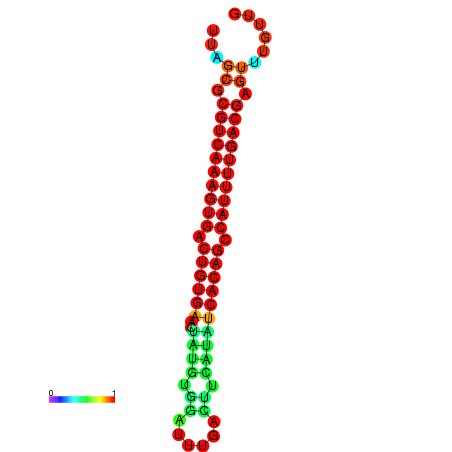 |
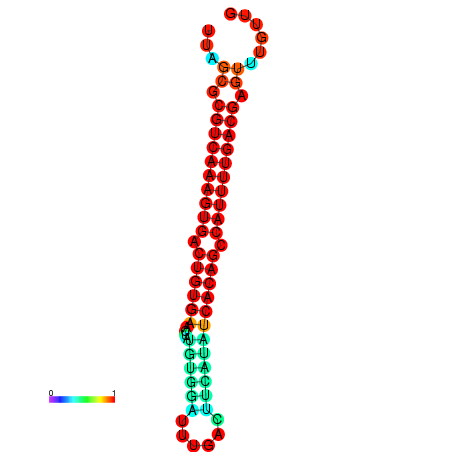 |
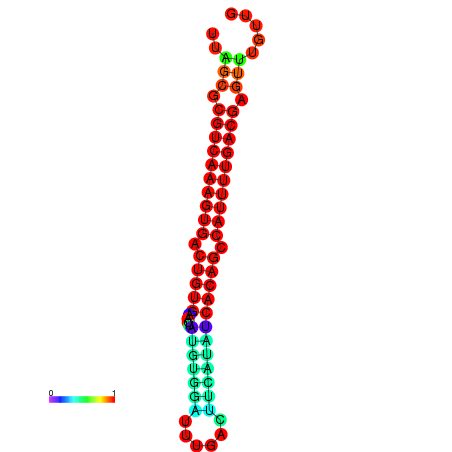 |
| dG=-29.0, p-value=0.009901 | dG=-29.0, p-value=0.009901 |
|---|---|
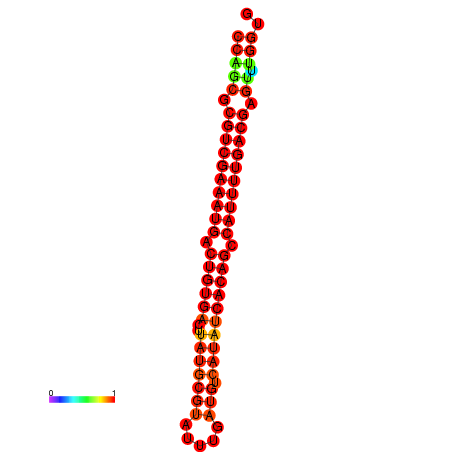 |
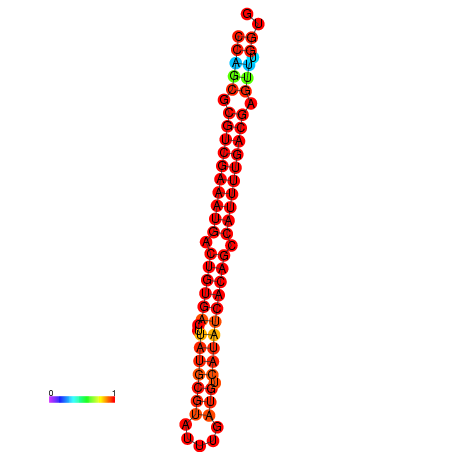 |
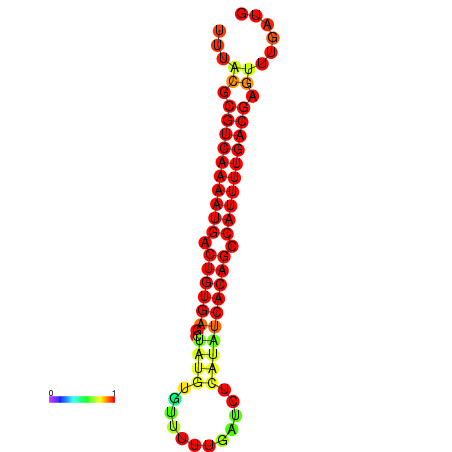
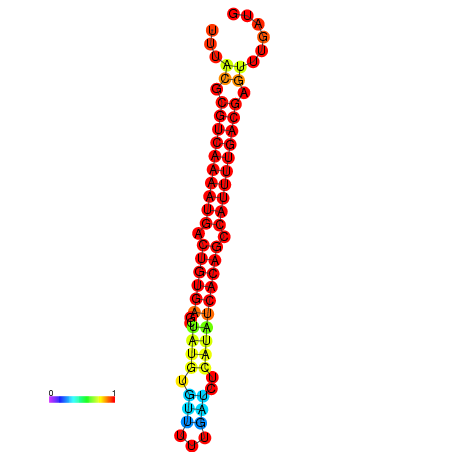
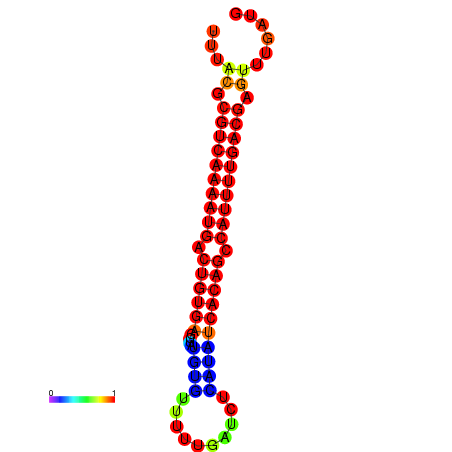
| dG=-23.6, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.0, p-value=0.009901 |
|---|---|---|
 |
 |
 |
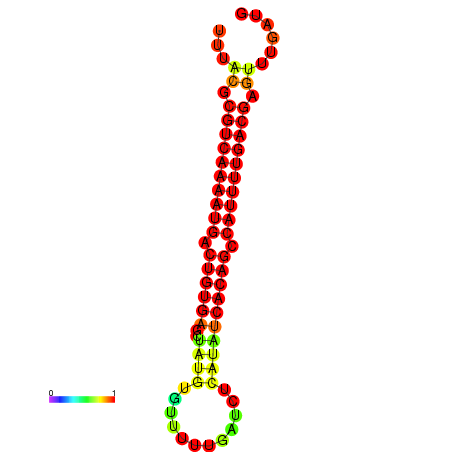
| dG=-23.6, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.0, p-value=0.009901 |
|---|---|---|
 |
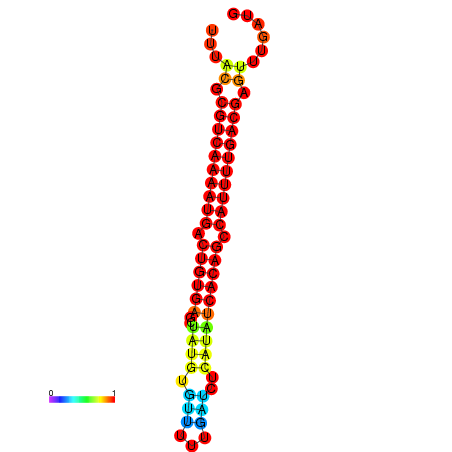 |
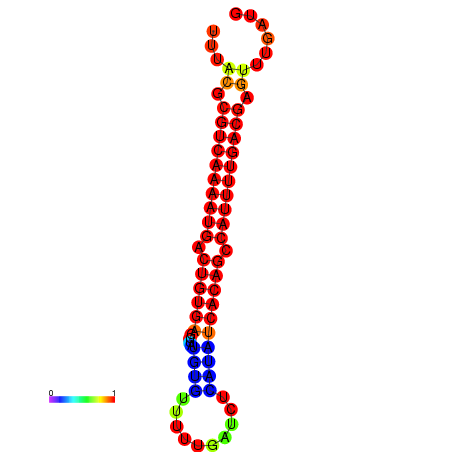 |
| dG=-23.5, p-value=0.009901 |
|---|
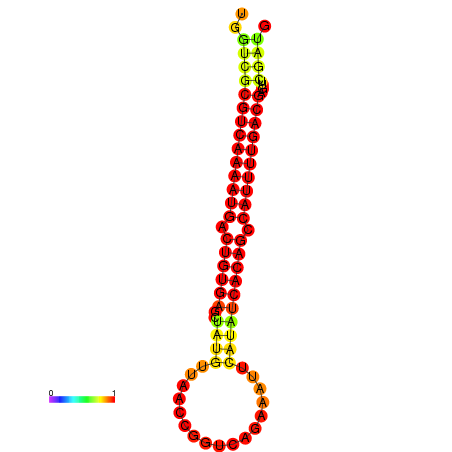 |
| dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|---|
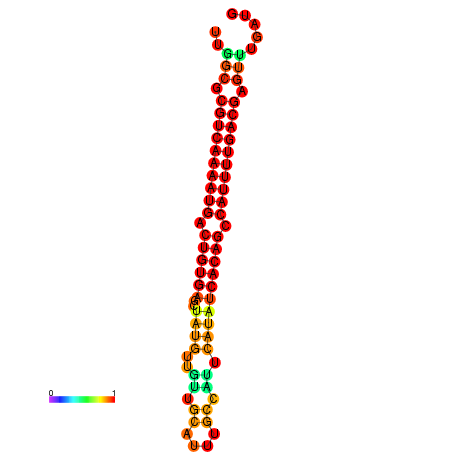 |
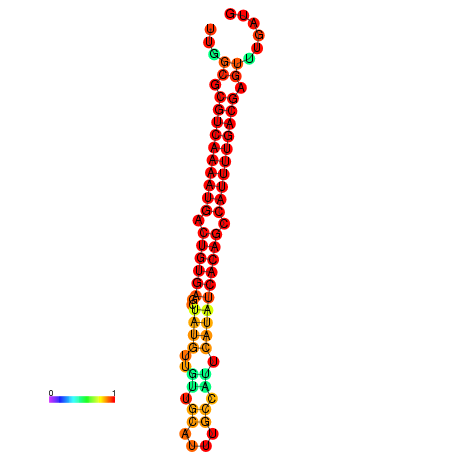 |
 |
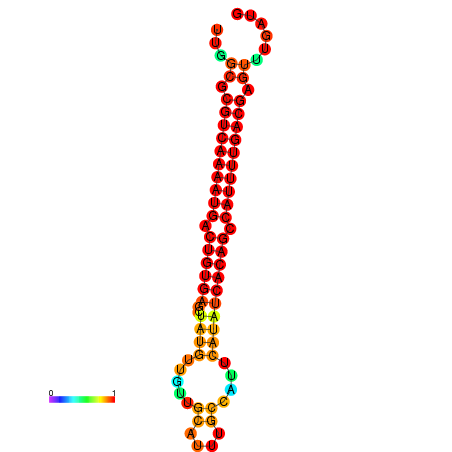 |
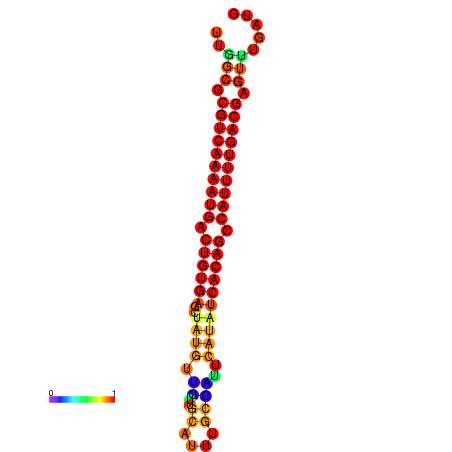 |
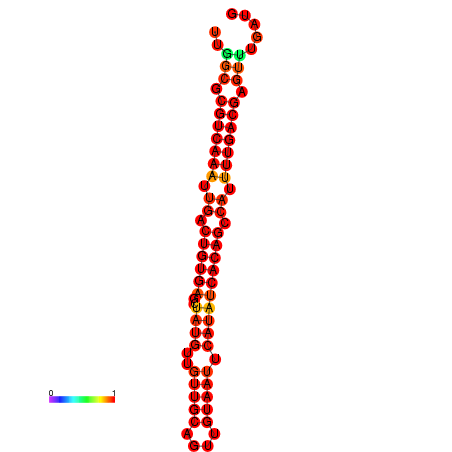
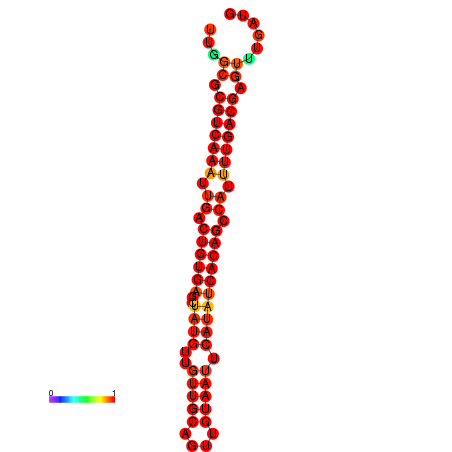
| dG=-21.6, p-value=0.009901 |
|---|
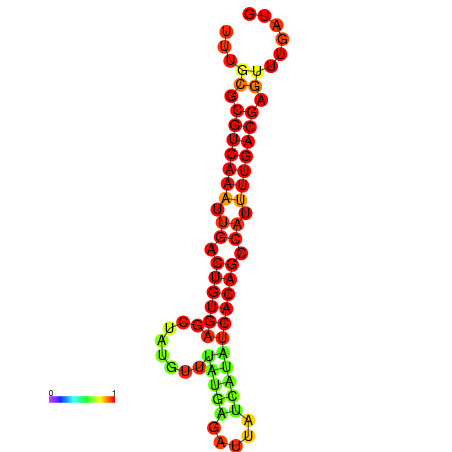 |
| dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|
 |
 |
Generated: 03/07/2013 at 04:51 PM