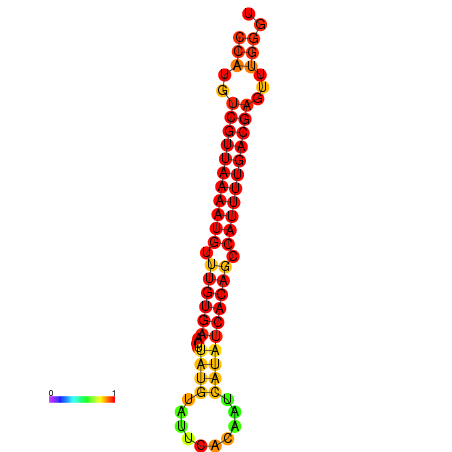


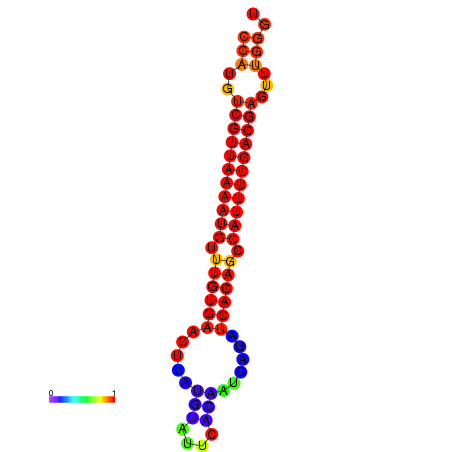
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:11243115-11243212 - | CACG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-ACAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droSim1 | chr3R:10237959-10238056 + | CGCG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droSec1 | super_0:10705190-10705287 + | CGCG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droYak2 | chr3R:15188722-15188819 + | CACG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droEre2 | scaffold_4770:10769258-10769355 - | AACG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droAna3 | scaffold_13340:12827469-12827566 - | --CA-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAAGGCTTGTGACCTTATGT---ATTTCGCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAC |
| dp4 | chr2:9825076-9825174 + | AGCA-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGACCTTATGT---ACTCTTGAATCATATCACAGCCATTTTGACGAGTTTGGGTACAGAAATTTCAA |
| droPer1 | super_0:10659921-10660019 - | AGCA-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGACCTTATGT---ACTCTTGAATCATATCACAGCCATTTTGACGAGTTTGGGTACAGAAATTTCAA |
| droWil1 | scaffold_181089:2143062-2143162 - | TGCA-----------------------------------------------------ACTTTCTTATACCATGTCGTTAAAATGTTTGTGACTTTATGTTGTGTCT-CTATTCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droVir3 | scaffold_13047:13335778-13335875 + | TGCA-----------------------------------------------------ATTTTCTTATACCATGTCGTTAAAATGCCTGTGACCTTATGT---ATTT-TATATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droMoj3 | scaffold_6540:10568509-10568606 + | TGCA-----------------------------------------------------ATTTTCTTATACCATGTCGTTAAAATGTCTGTGACCGTATGT---ATTC-ACTATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droGri2 | scaffold_14906:4357489-4357639 - | CGTGCAGTCGCCGCCTCCTCCTGTTCCTCCTCCTTCTCCTTTGTGTGTTTCTTTCTTTCTTTCTTATACCATGTCGTTAAAATGCTTGTGACCTTATGT---ATTT-TATATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATGTCAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:11243115-11243212 - | CACG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-ACAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droSim1 | chr3R:10237959-10238056 + | CGCG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droSec1 | super_0:10705190-10705287 + | CGCG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droYak2 | chr3R:15188722-15188819 + | CACG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droEre2 | scaffold_4770:10769258-10769355 - | AACG-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGAACTTATGT---ATTC-GCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droAna3 | scaffold_13340:12827469-12827566 - | --CA-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAAGGCTTGTGACCTTATGT---ATTTCGCAATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAC |
| dp4 | chr2:9825076-9825174 + | AGCA-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGACCTTATGT---ACTCTTGAATCATATCACAGCCATTTTGACGAGTTTGGGTACAGAAATTTCAA |
| droPer1 | super_0:10659921-10660019 - | AGCA-----------------------------------------------------TCTTTCTTATACCATGTCGTTAAAATGTTTGTGACCTTATGT---ACTCTTGAATCATATCACAGCCATTTTGACGAGTTTGGGTACAGAAATTTCAA |
| droWil1 | scaffold_181089:2143062-2143162 - | TGCA-----------------------------------------------------ACTTTCTTATACCATGTCGTTAAAATGTTTGTGACTTTATGTTGTGTCT-CTATTCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droVir3 | scaffold_13047:13335778-13335875 + | TGCA-----------------------------------------------------ATTTTCTTATACCATGTCGTTAAAATGCCTGTGACCTTATGT---ATTT-TATATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droMoj3 | scaffold_6540:10568509-10568606 + | TGCA-----------------------------------------------------ATTTTCTTATACCATGTCGTTAAAATGTCTGTGACCGTATGT---ATTC-ACTATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATCTCAA |
| droGri2 | scaffold_14906:4357489-4357639 - | CGTGCAGTCGCCGCCTCCTCCTGTTCCTCCTCCTTCTCCTTTGTGTGTTTCTTTCTTTCTTTCTTATACCATGTCGTTAAAATGCTTGTGACCTTATGT---ATTT-TATATCATATCACAGCCATTTTGACGAGTTTGGGTAAAGAAATGTCAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-21.9, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-22.0, p-value=0.009901 |
|---|---|---|---|
 |
 |
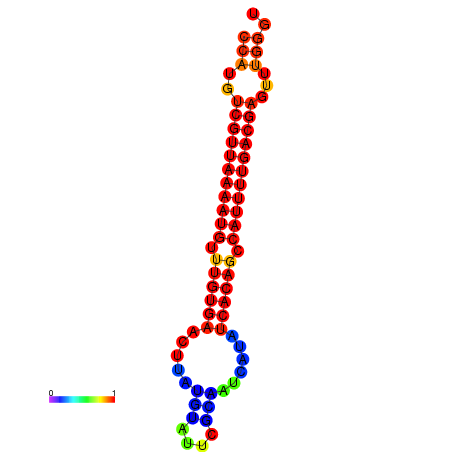 |
 |
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-22.0, p-value=0.009901 |
|---|---|---|---|
 |
 |
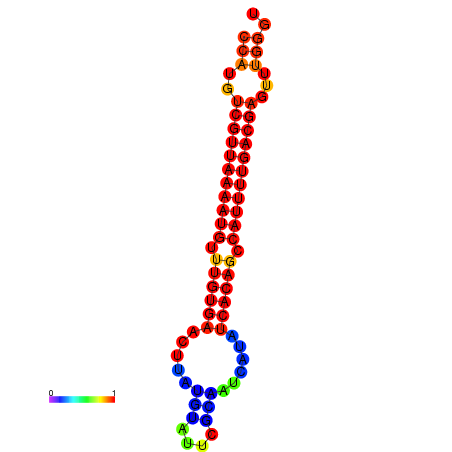 |
 |
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-22.0, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-22.0, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
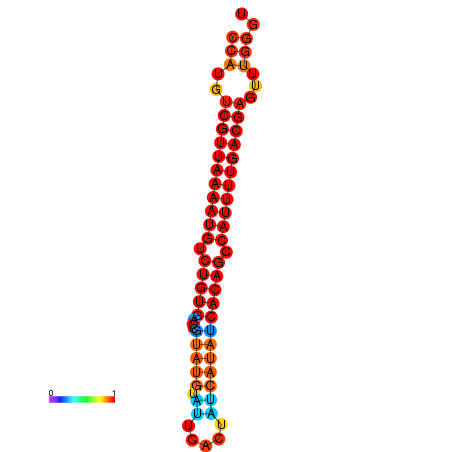
| dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
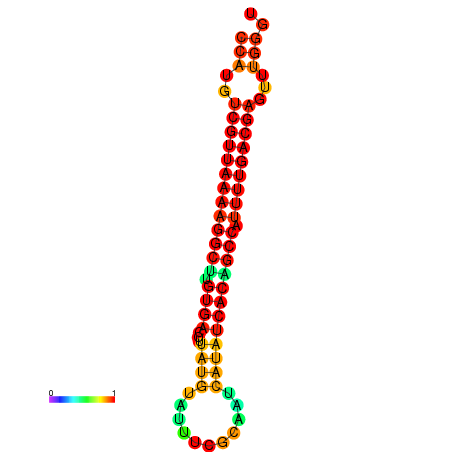 |
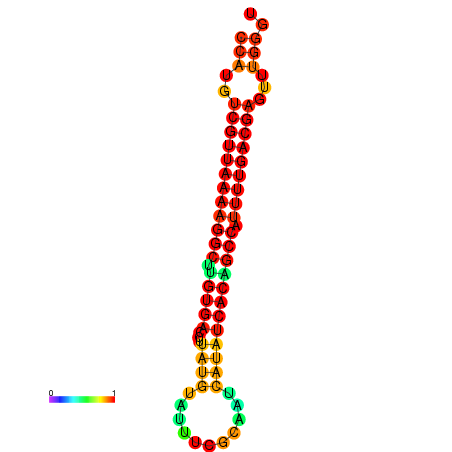 |
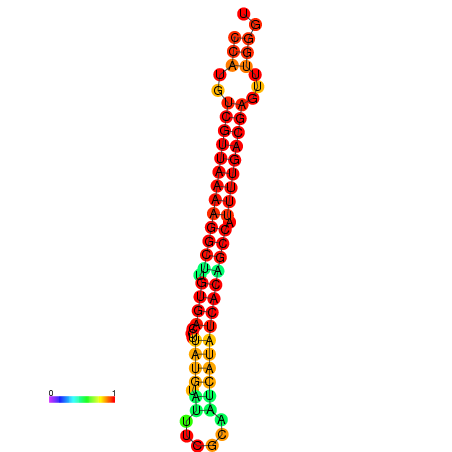 |
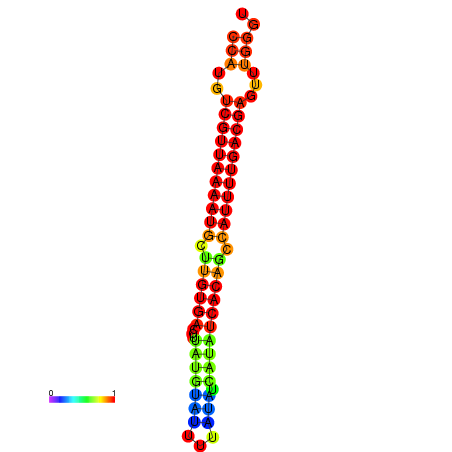
| dG=-22.8, p-value=0.009901 |
|---|
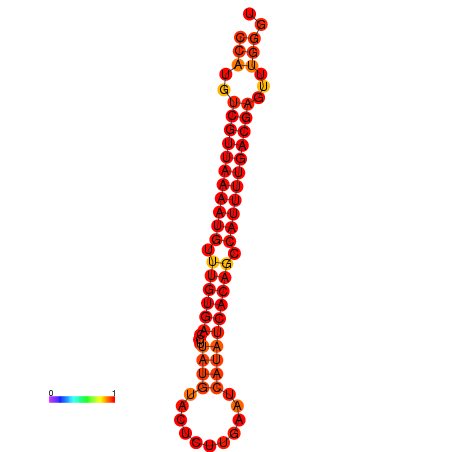 |
| dG=-22.8, p-value=0.009901 |
|---|
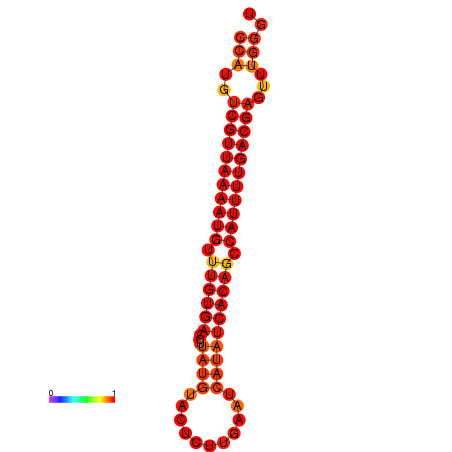 |
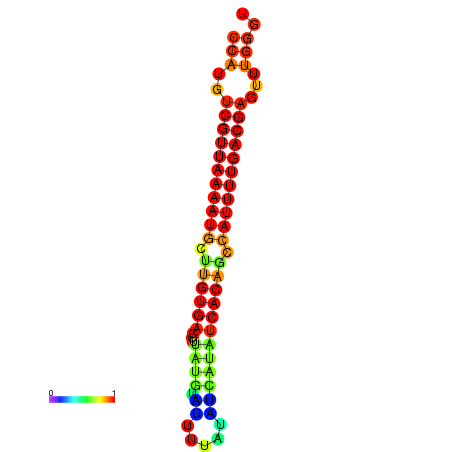
| dG=-22.7, p-value=0.009901 | dG=-22.6, p-value=0.009901 | dG=-22.4, p-value=0.009901 | dG=-21.9, p-value=0.009901 |
|---|---|---|---|
 |
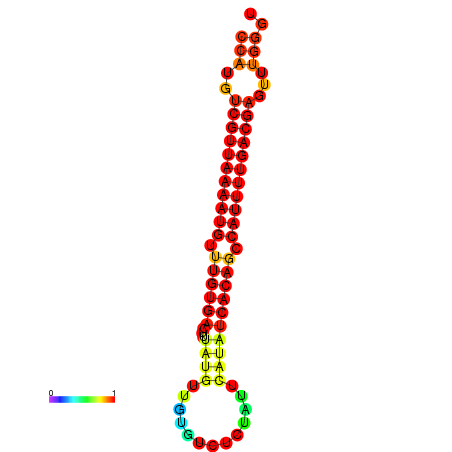 |
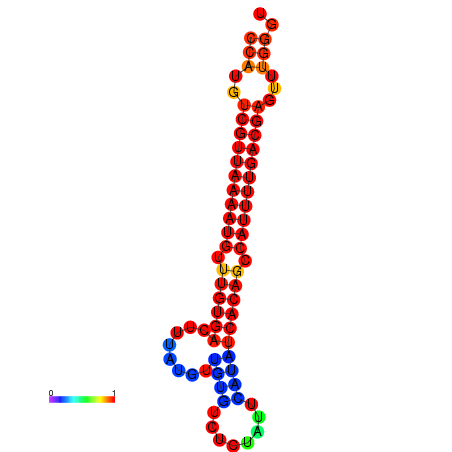 |
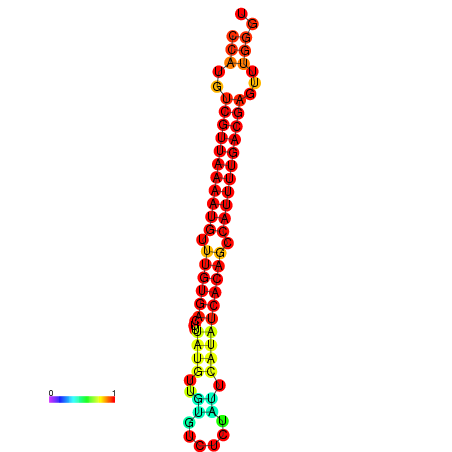 |
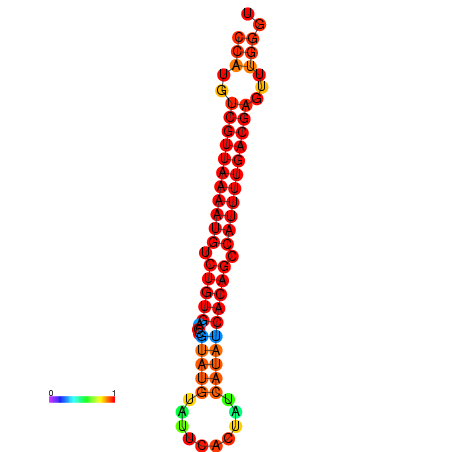
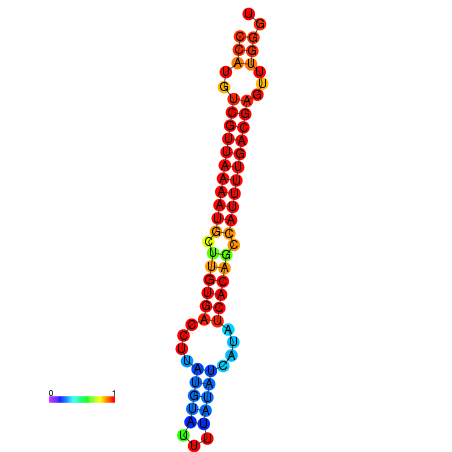
| dG=-25.1, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.1, p-value=0.009901 |
|---|---|---|---|---|
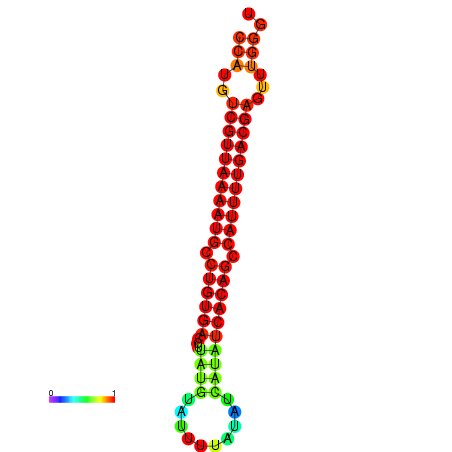 |
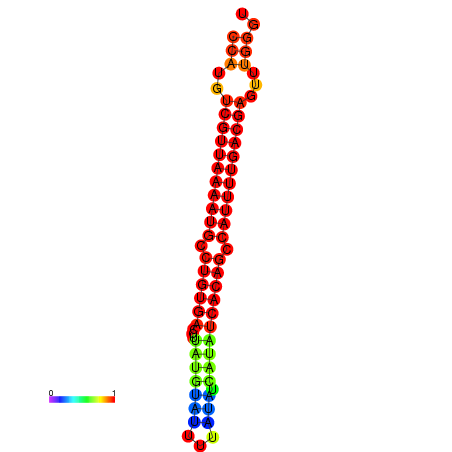 |
 |
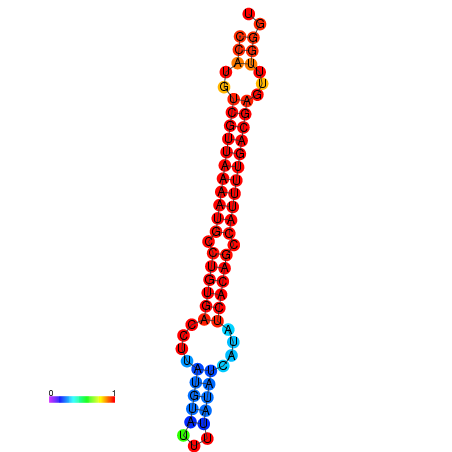 |
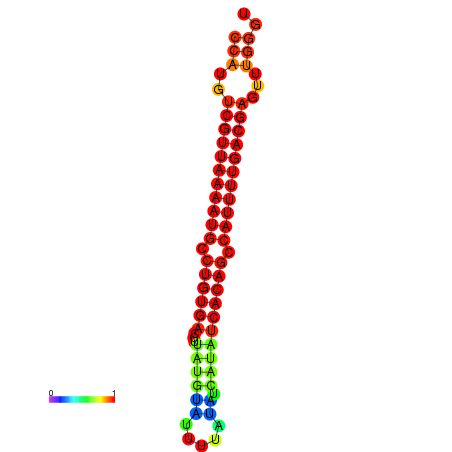 |
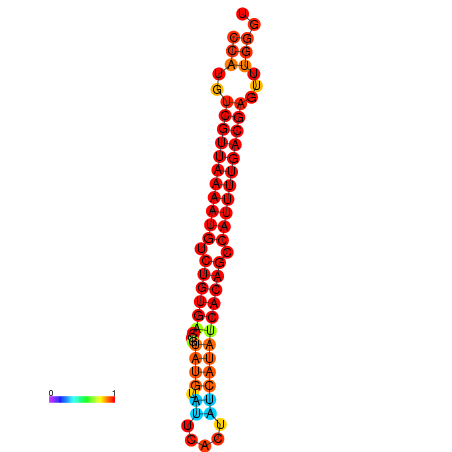
| dG=-25.1, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.4, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
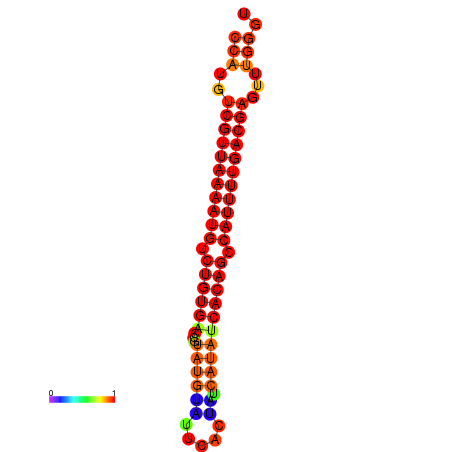 |
| dG=-22.9, p-value=0.009901 | dG=-22.7, p-value=0.009901 | dG=-22.7, p-value=0.009901 | dG=-22.7, p-value=0.009901 | dG=-21.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:51 PM