| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:11243249-11243353 - | CCAAG----C------------------CAT--AATTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--GCG----GCA----------------------GCG |
| droSim1 | chr3R:10237822-10237926 + | CCAAG----C------------------CAT--AGTTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--TCG----GCA----------------------GCG |
| droSec1 | super_0:10705053-10705157 + | CCAAG----C------------------CAT--AGTTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--TGG----TCA----------------------GCG |
| droYak2 | chr3R:15188577-15188681 + | CCAAG----C------------------CAT--AATTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGCC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--TCG----GCA----------------------ACT |
| droEre2 | scaffold_4770:10769389-10769493 - | CCAAG----A------------------CAC--AATTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-T--TAA----AAA----------------------GCG |
| droAna3 | scaffold_13340:12827611-12827739 - | ACAAGAATCCCCTCAAAAAAGGACGATCCATCGAGTTCTTACGTAACTCCTCAAAG-GGCTGTGAAATGTTGCCA--TTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGGGA-AT-C-GTCA----GAA----------------------AAC |
| dp4 | chr2:9824930-9825029 + | CC-------C------------------CA-----TACTTACGTAACCCATCAAAC-GGTTGTGAAATGTTGCAT--TTA-TCAAATCATATCACAGCCATTTTGATGAGTTTCGTGTGG-AA-TGGCCA----GCA----------------------ATC |
| droPer1 | super_0:10660066-10660165 - | CC-------C------------------CA-----TACTTACGTAACCCATCAAAC-GGTTGTGAAATGTTGCAT--TTA-TCAAATCATATCACAGCCATTTTGATGAGTTTCGTGTGG-AA-TGGCCA----GCA----------------------ATC |
| droWil1 | scaffold_181089:2143204-2143324 - | CCGTT----CCGGCAAAAGATG-------GA--ATTTCTTACGTAACTCTTCAAGGGGGTTGTGAAATGTTGCCATTTTATTCAACTCATATCACAGCCATTTTGATGAGTTTCGTGCGAGTATT--CCT----GCA----------------------ACA |
| droVir3 | scaffold_13047:13335625-13335725 + | CCACT----C-----------------------AATTCTTGCGTAACTCTTCAAGG-GGTTGTGAAATGTTGCCG--TTA-TCAAGTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-T--GCT----TAA----------------------ATT |
| droMoj3 | scaffold_6540:10568336-10568468 + | CTCCG----C-----------------CCATTCAATTCTTGCGTAACTCTTCAAGG-GGTTGTGAAATGTAGCCA--TTA-TCAAGTCATATCACAGCCATTTTGATGAGTTTCGTGTGA-AT-T--GCTTTCAGCATTGTCCCATCCGTTTTGTGTCACTG |
| droGri2 | scaffold_14906:4357674-4357778 - | CCAAG----C-----------------CCACCCAATTCTTGCGTAACTCTTCAAGG-GGTTGTGAAATGTGGGTA--TTA-TCAAGTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-T--GCTT---T-------------------------CA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:11243249-11243353 - | CCAAG----C------------------CAT--AATTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--GCG----GCA----------------------GCG |
| droSim1 | chr3R:10237822-10237926 + | CCAAG----C------------------CAT--AGTTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--TCG----GCA----------------------GCG |
| droSec1 | super_0:10705053-10705157 + | CCAAG----C------------------CAT--AGTTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--TGG----TCA----------------------GCG |
| droYak2 | chr3R:15188577-15188681 + | CCAAG----C------------------CAT--AATTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGCC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-C--TCG----GCA----------------------ACT |
| droEre2 | scaffold_4770:10769389-10769493 - | CCAAG----A------------------CAC--AATTCTTACGTAACTCCTCAAAG-GGTTGTGAAATGTCGAC-TATTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-T--TAA----AAA----------------------GCG |
| droAna3 | scaffold_13340:12827611-12827739 - | ACAAGAATCCCCTCAAAAAAGGACGATCCATCGAGTTCTTACGTAACTCCTCAAAG-GGCTGTGAAATGTTGCCA--TTA-TCTACTCATATCACAGCCATTTTGATGAGTTTCGTGGGA-AT-C-GTCA----GAA----------------------AAC |
| dp4 | chr2:9824930-9825029 + | CC-------C------------------CA-----TACTTACGTAACCCATCAAAC-GGTTGTGAAATGTTGCAT--TTA-TCAAATCATATCACAGCCATTTTGATGAGTTTCGTGTGG-AA-TGGCCA----GCA----------------------ATC |
| droPer1 | super_0:10660066-10660165 - | CC-------C------------------CA-----TACTTACGTAACCCATCAAAC-GGTTGTGAAATGTTGCAT--TTA-TCAAATCATATCACAGCCATTTTGATGAGTTTCGTGTGG-AA-TGGCCA----GCA----------------------ATC |
| droWil1 | scaffold_181089:2143204-2143324 - | CCGTT----CCGGCAAAAGATG-------GA--ATTTCTTACGTAACTCTTCAAGGGGGTTGTGAAATGTTGCCATTTTATTCAACTCATATCACAGCCATTTTGATGAGTTTCGTGCGAGTATT--CCT----GCA----------------------ACA |
| droVir3 | scaffold_13047:13335625-13335725 + | CCACT----C-----------------------AATTCTTGCGTAACTCTTCAAGG-GGTTGTGAAATGTTGCCG--TTA-TCAAGTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-T--GCT----TAA----------------------ATT |
| droMoj3 | scaffold_6540:10568336-10568468 + | CTCCG----C-----------------CCATTCAATTCTTGCGTAACTCTTCAAGG-GGTTGTGAAATGTAGCCA--TTA-TCAAGTCATATCACAGCCATTTTGATGAGTTTCGTGTGA-AT-T--GCTTTCAGCATTGTCCCATCCGTTTTGTGTCACTG |
| droGri2 | scaffold_14906:4357674-4357778 - | CCAAG----C-----------------CCACCCAATTCTTGCGTAACTCTTCAAGG-GGTTGTGAAATGTGGGTA--TTA-TCAAGTCATATCACAGCCATTTTGATGAGTTTCGTGCGA-AT-T--GCTT---T-------------------------CA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
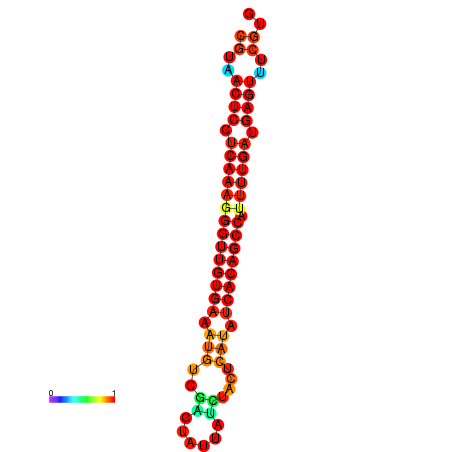 |
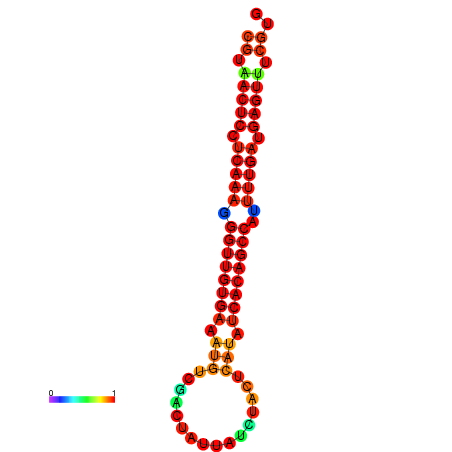 |
| dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|---|---|
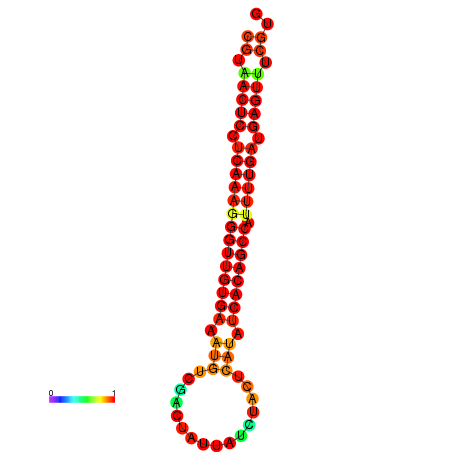 |
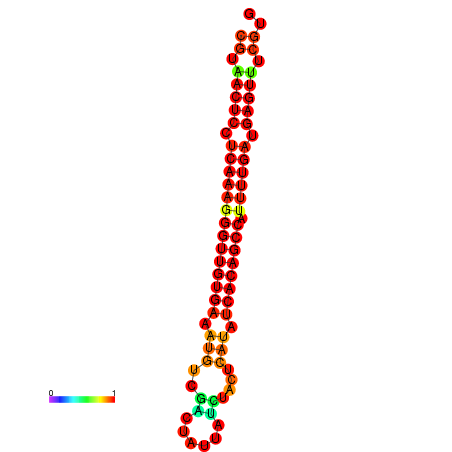 |
 |
 |
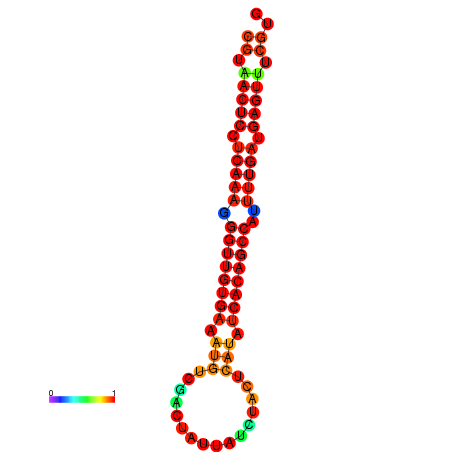 |
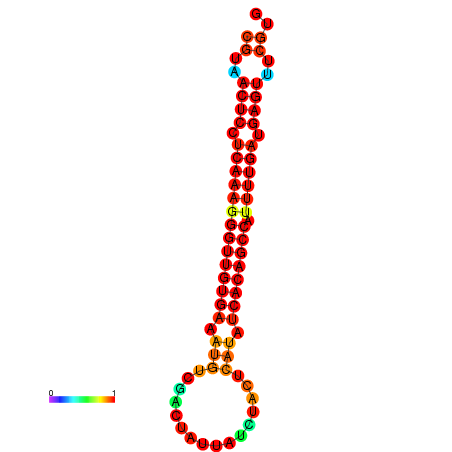
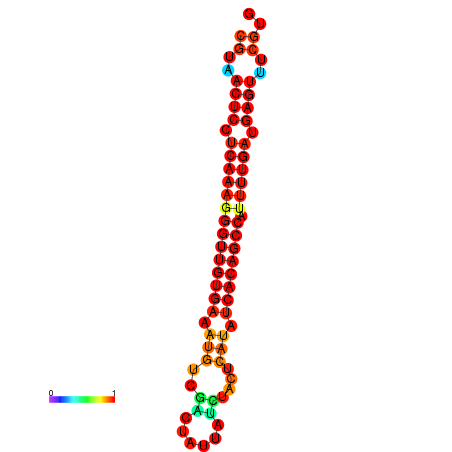
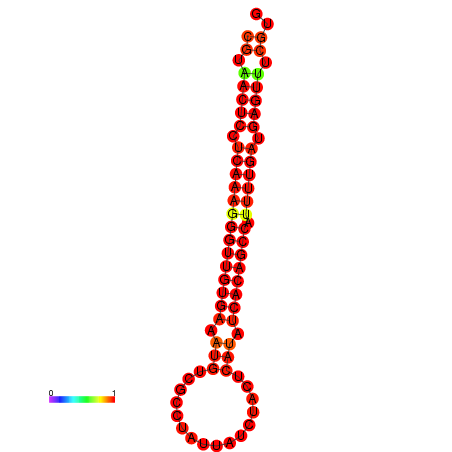
| dG=-25.1, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
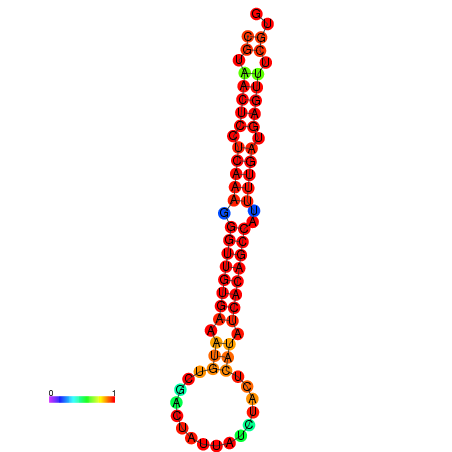 |
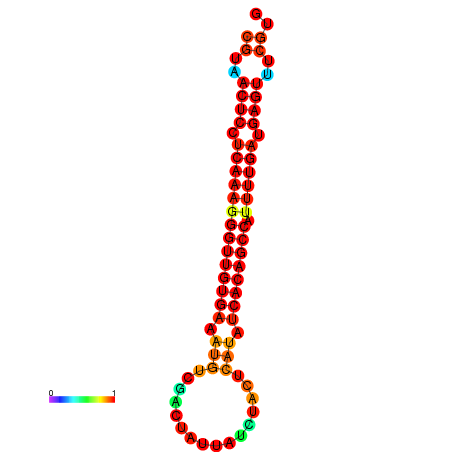
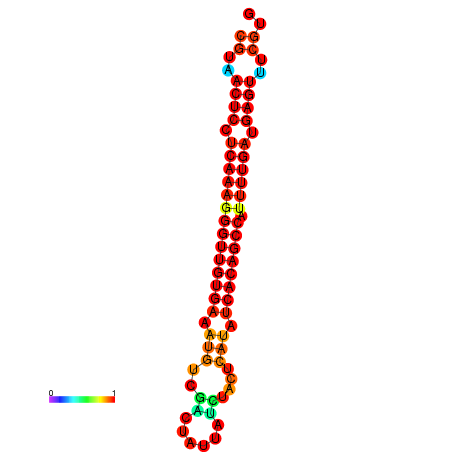
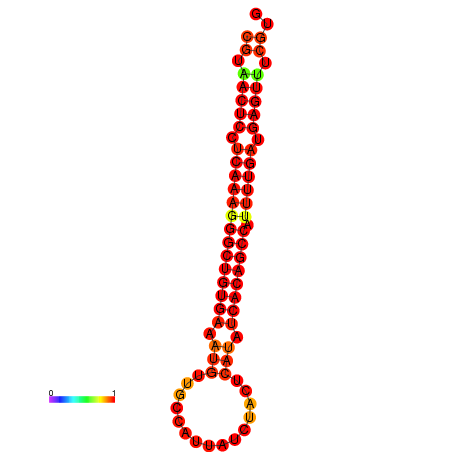
| dG=-27.6, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-26.8, p-value=0.009901 |
|---|---|---|
 |
 |
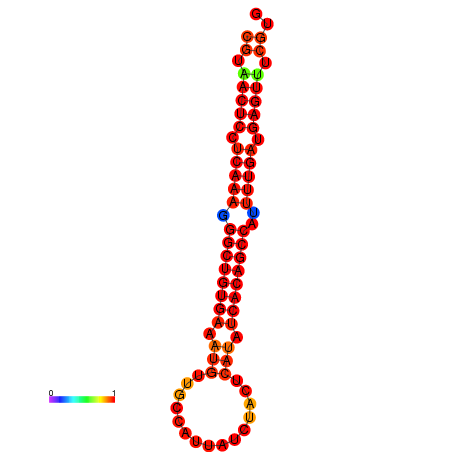 |
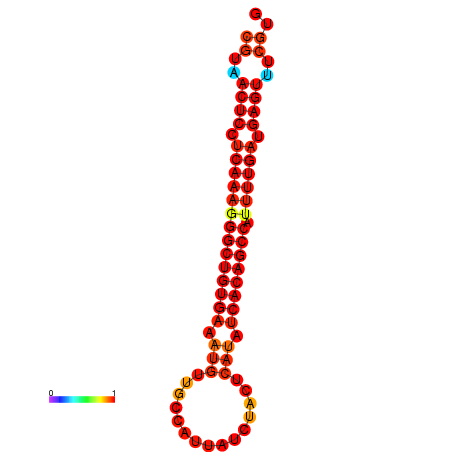
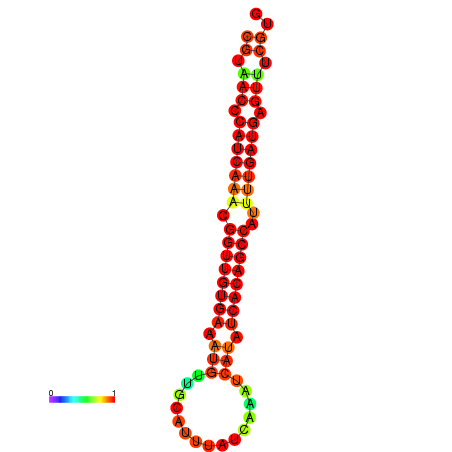
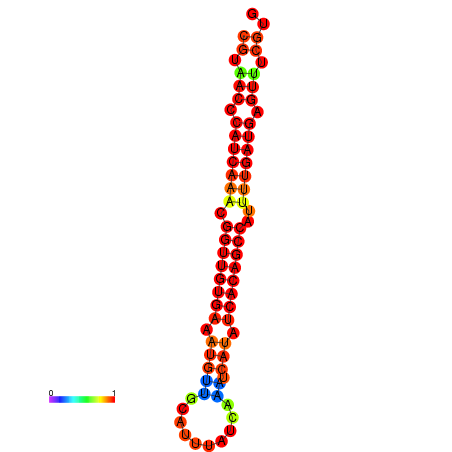
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.4, p-value=0.009901 | dG=-22.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
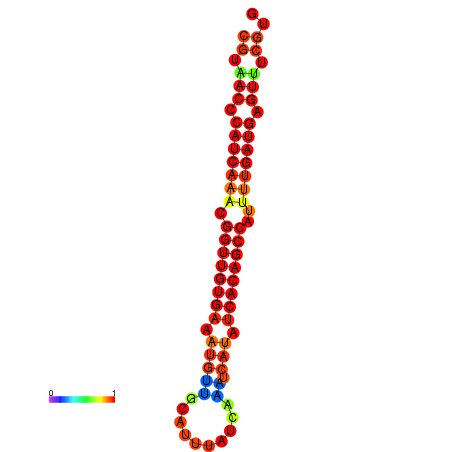
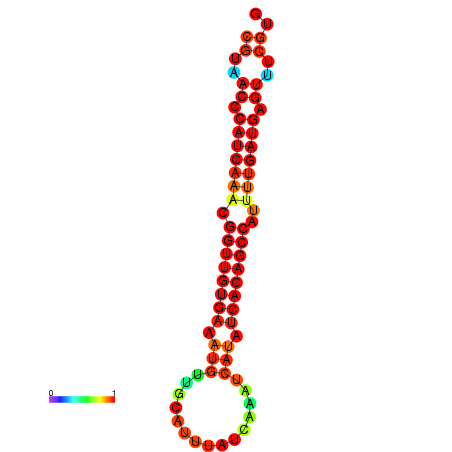
| dG=-22.9, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.4, p-value=0.009901 | dG=-22.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
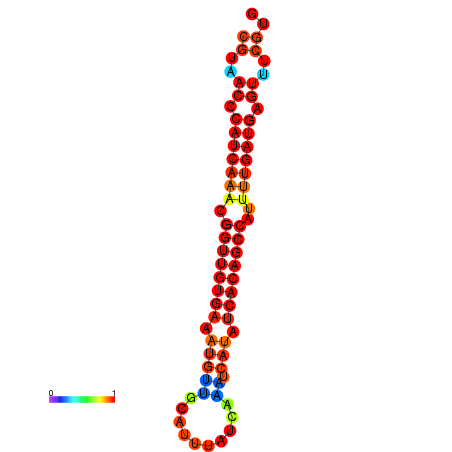 |
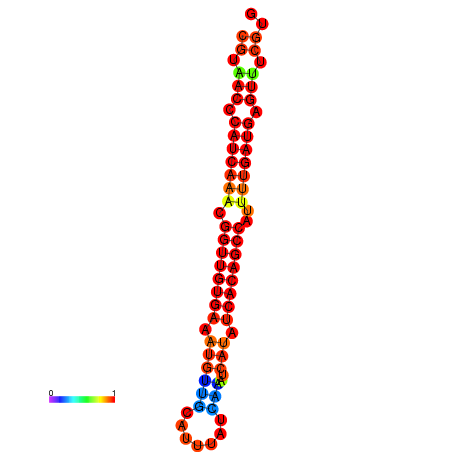
| dG=-25.5, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
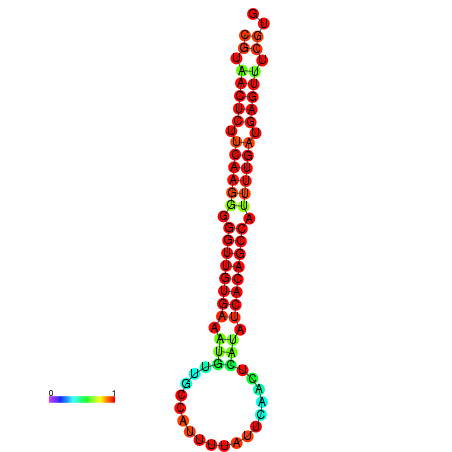 |
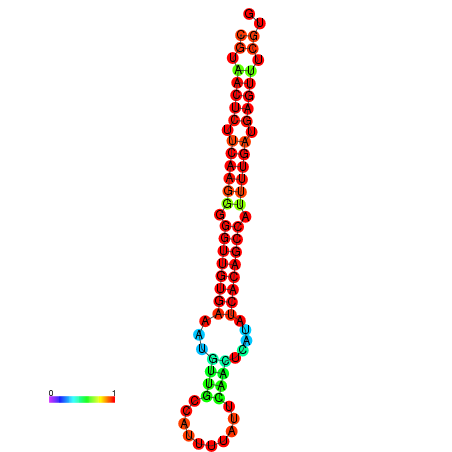 |
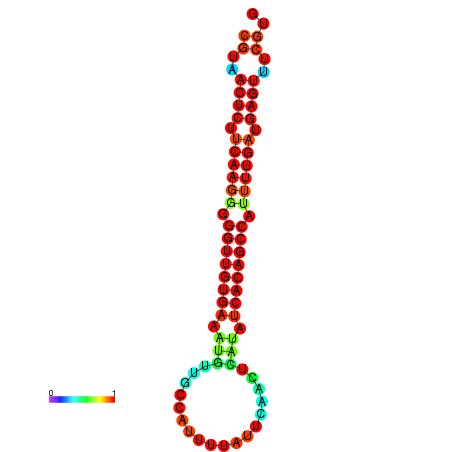 |
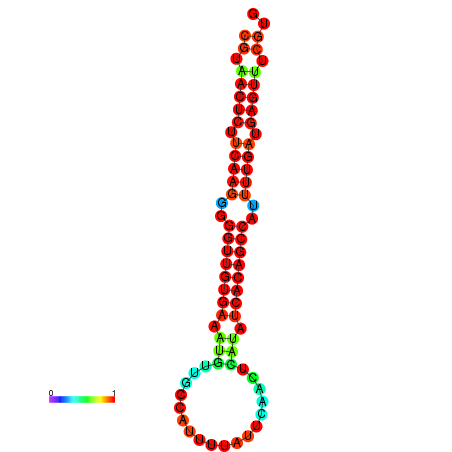 |
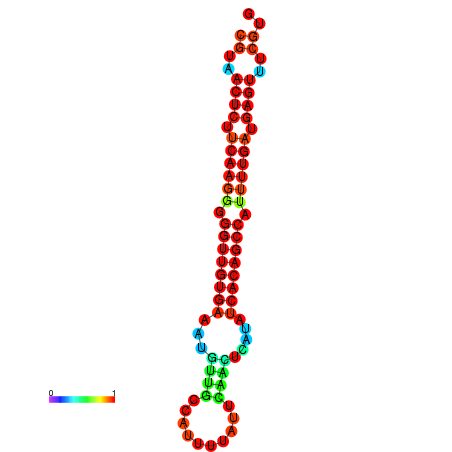 |
| dG=-25.2, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|---|
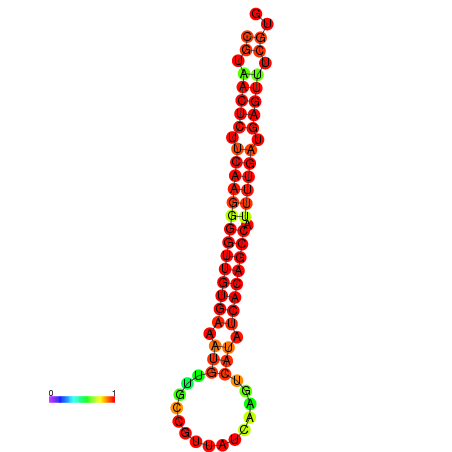 |
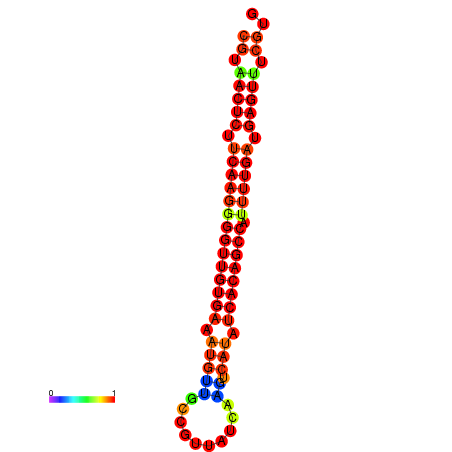 |
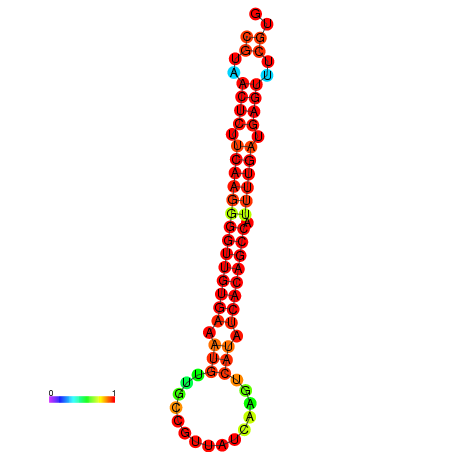 |
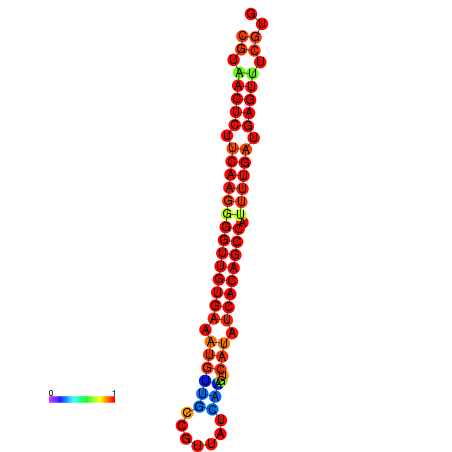 |
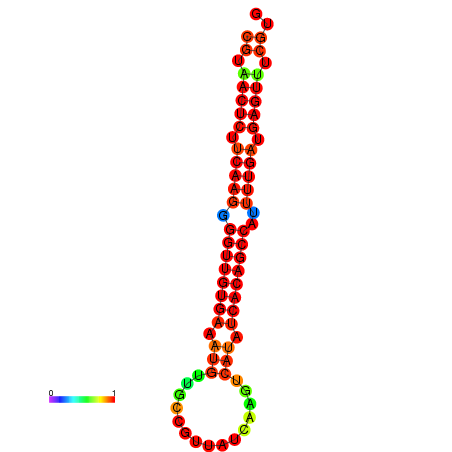 |
| dG=-25.2, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|---|
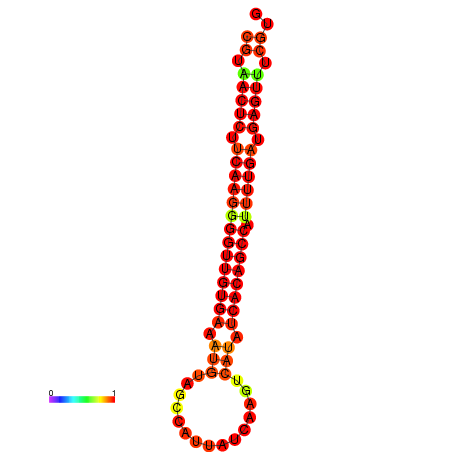 |
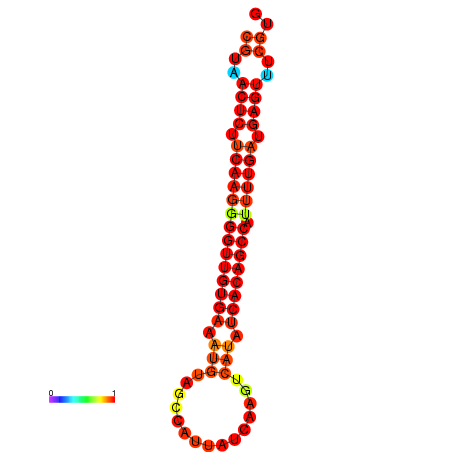 |
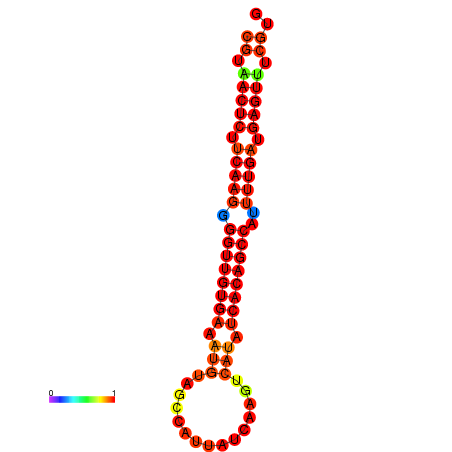 |
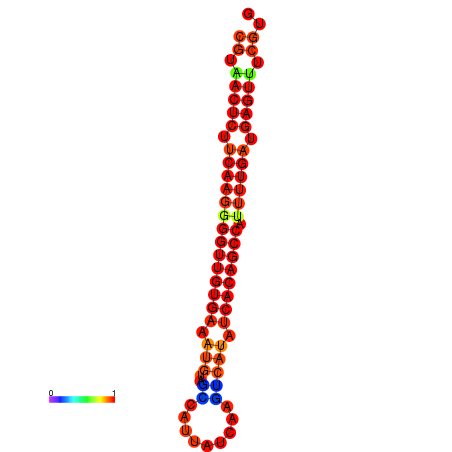 |
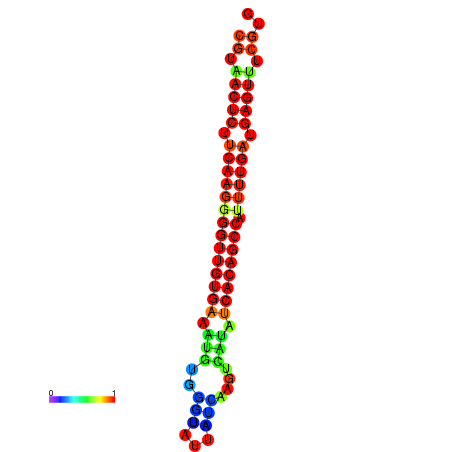
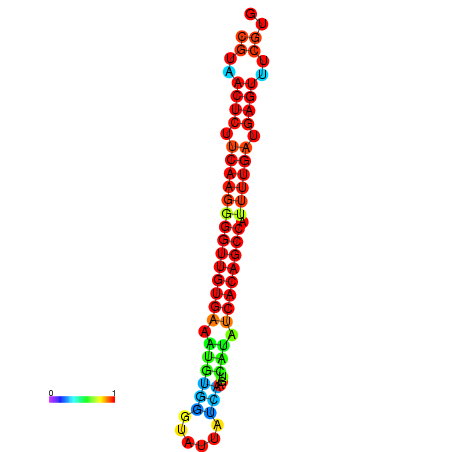
| dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
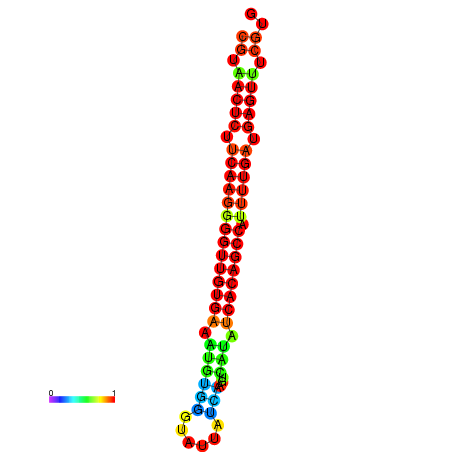 |
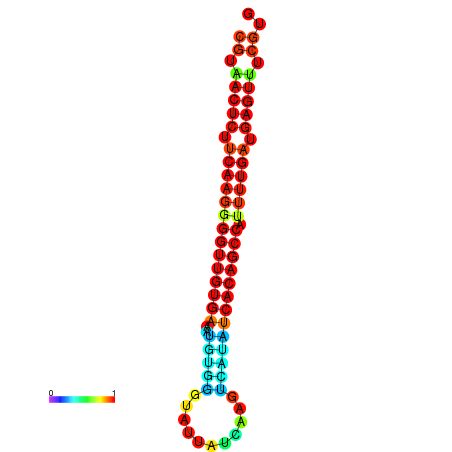 |
 |
 |
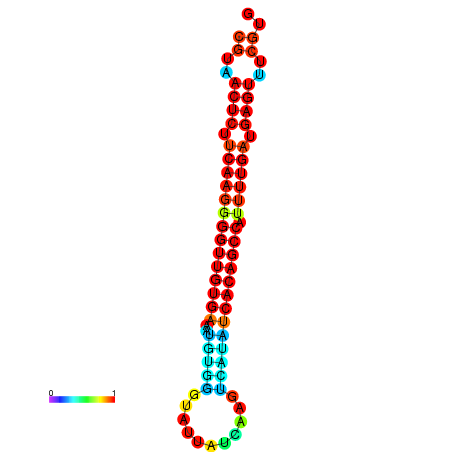 |
Generated: 03/07/2013 at 04:51 PM