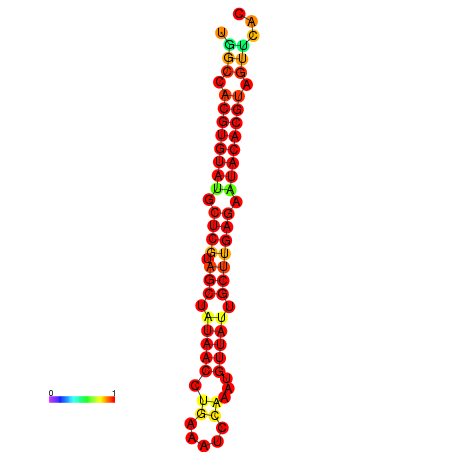
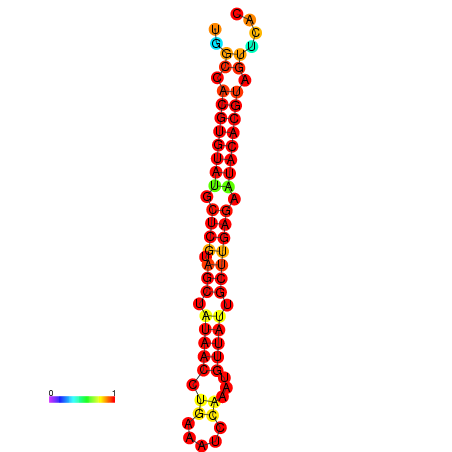
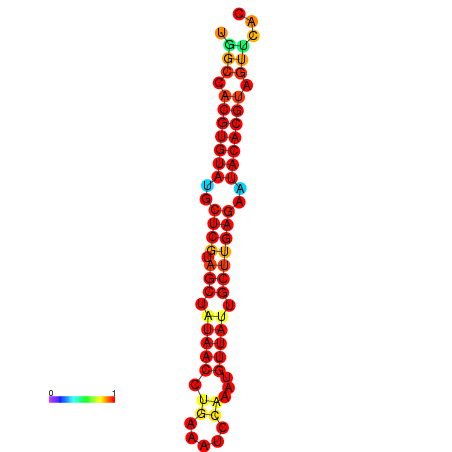
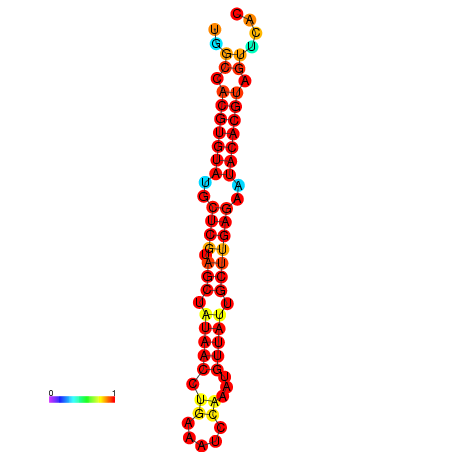
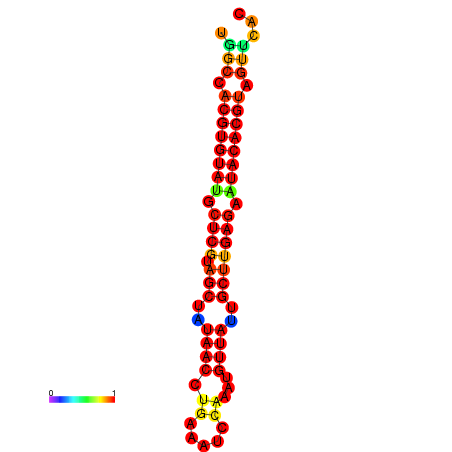
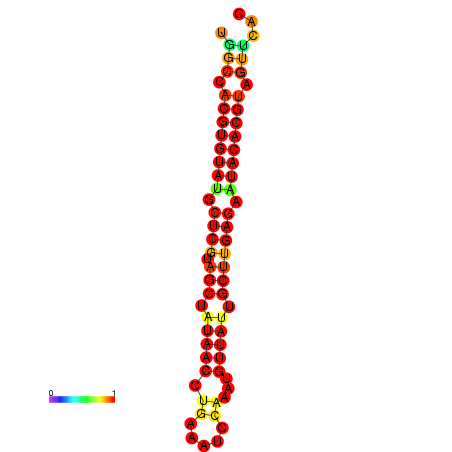
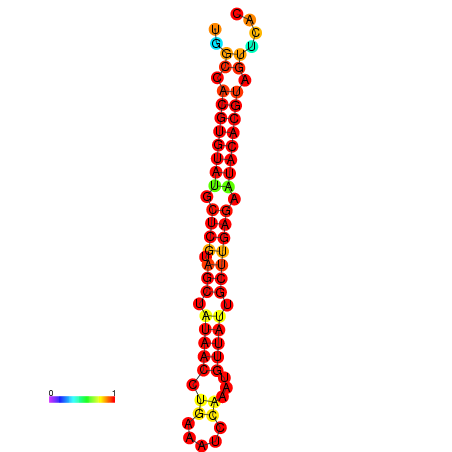
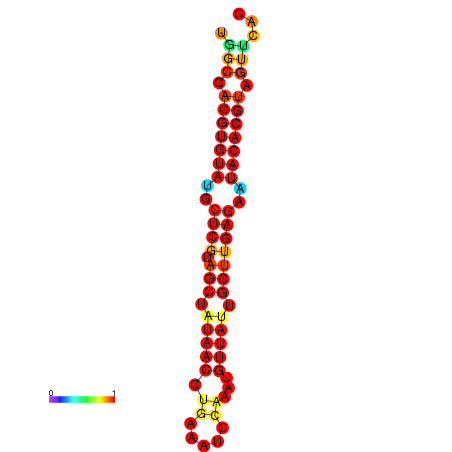
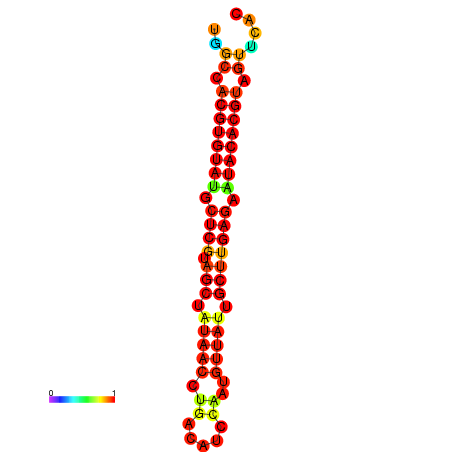
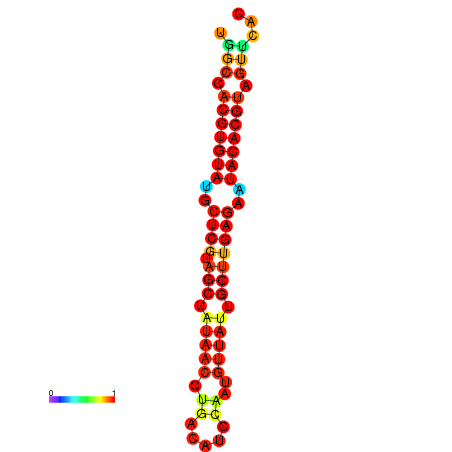
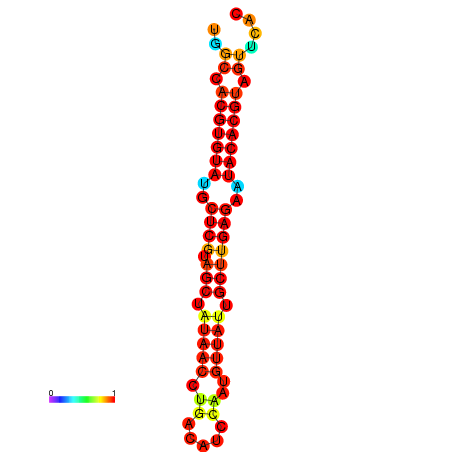
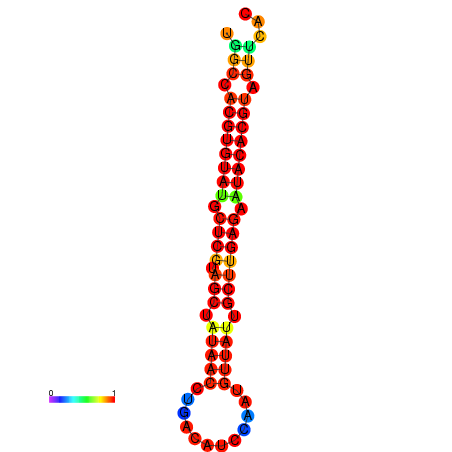
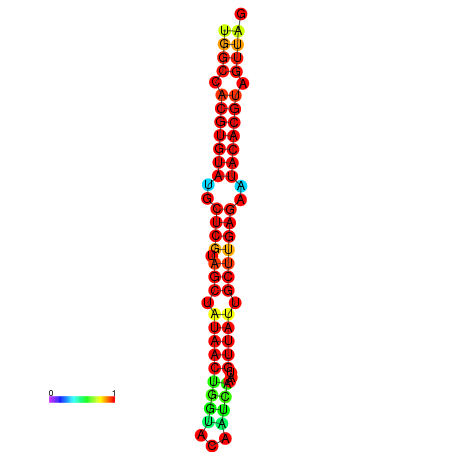
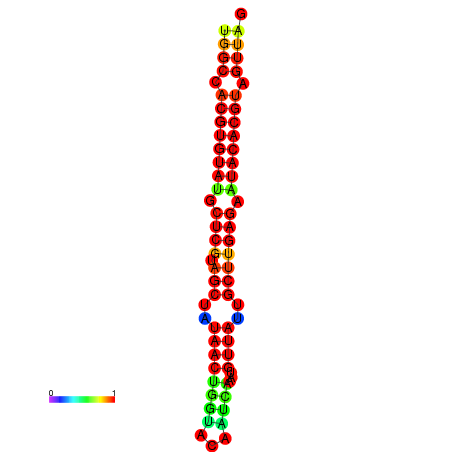
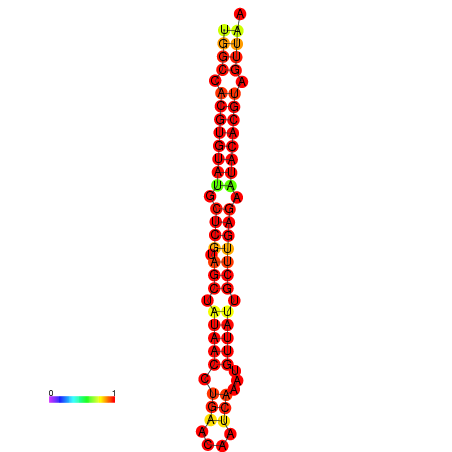
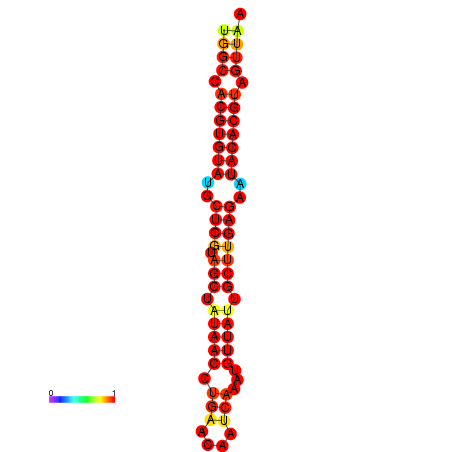
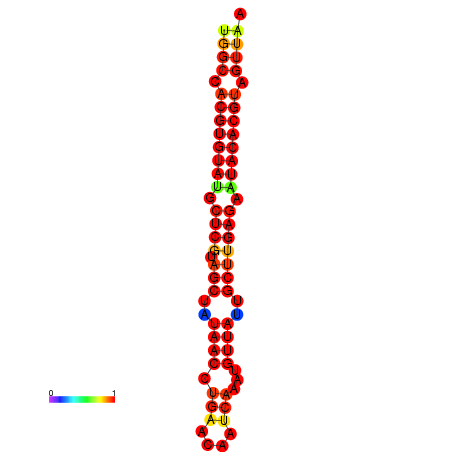
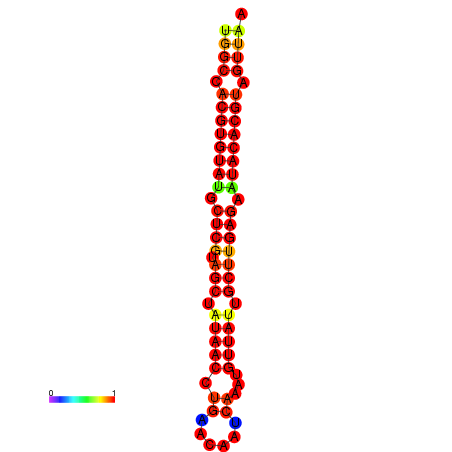
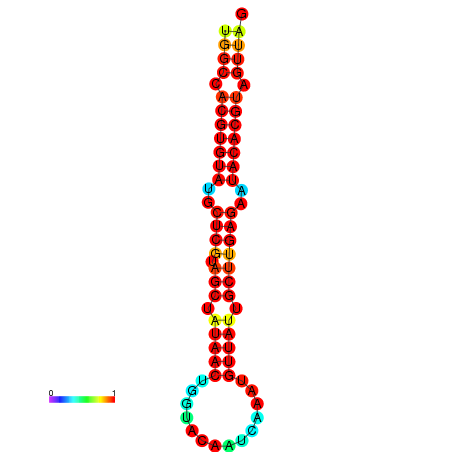| TTAAGATGACAGCGACGGCCAAGGCAATGCCGACGGCGACTAGAATCTTCAACGGCCACGTGTATACTCATAGCTATAACCTGATATCCGAATGTTATTGCTTGAGAATACACGTAGTTTAACAAGATTCGTTCAGCAATCAAACTGAAACGGAAAGTGTCTGTGAAAGGTGTCCTTAAAG | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ...............................................................................................TATTGCTTGAGAATACACGTAG................................................................ | 22 | 1 | 1204.00 | 1204 | 437 | 767 |
| ...............................................................................................TATTGCTTGAGAATACACGTA................................................................. | 21 | 1 | 139.00 | 139 | 14 | 125 |
| .........................................................ACGTGTATACTCATAGCTATAAC..................................................................................................... | 23 | 1 | 87.00 | 87 | 82 | 5 |
| ...............................................................................................TATTGCTTGAGAATACACGT.................................................................. | 20 | 1 | 61.00 | 61 | 1 | 60 |
| ...............................................................................................TATTGCTTGAGAATACACGTAGT............................................................... | 23 | 1 | 27.00 | 27 | 11 | 16 |
| ................................................................................................ATTGCTTGAGAATACACGTAGT............................................................... | 22 | 1 | 19.00 | 19 | 5 | 14 |
| ................................................................................................ATTGCTTGAGAATACACGTA................................................................. | 20 | 1 | 14.00 | 14 | 0 | 14 |
| ........................................................CACGTGTATACTCATAGCTATAA...................................................................................................... | 23 | 1 | 10.00 | 10 | 8 | 2 |
| ................................................................................................ATTGCTTGAGAATACACGTAG................................................................ | 21 | 1 | 7.00 | 7 | 2 | 5 |
| ................................................................................................ATTGCTTGAGAATACACGT.................................................................. | 19 | 1 | 7.00 | 7 | 0 | 7 |
| .........................................................ACGTGTATACTCATAGCTATAA...................................................................................................... | 22 | 1 | 5.00 | 5 | 4 | 1 |
| .........................................................ACGTGTATACTCATAGCTATA....................................................................................................... | 21 | 1 | 5.00 | 5 | 3 | 2 |
| ........................................................CACGTGTATACTCATAGCTATA....................................................................................................... | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ...............................................................................................TATTGCTTGAGAATACACG................................................................... | 19 | 1 | 3.00 | 3 | 0 | 3 |
| .................................................................................................TTGCTTGAGAATACACGTAG................................................................ | 20 | 1 | 2.00 | 2 | 0 | 2 |
| ..............................................................................................TTATTGCTTGAGAATACACGT.................................................................. | 21 | 1 | 2.00 | 2 | 0 | 2 |
| .................................................................................................TTGCTTGAGAATACACGTAGTT.............................................................. | 22 | 1 | 2.00 | 2 | 1 | 1 |
| .................................................................................................TTGCTTGAGAATACACGTAGT............................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 |
| .........................................................ACGTGTATACTCATAGCTAT........................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 1 |
| ...................................................................................................GCTTGAGAATACACGTAG................................................................ | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ................................................................................................ATTGCTTGAGAATACACGTAGTT.............................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 |
| .........................................................ACGTGTATACTCATAGCTA......................................................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 |