| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:20616602-20616729 + | CAT--------TCGAGAA---------------------------TTTCCACC--------TGCAAC-ACTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-TCAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGATGACAACGAAAG-CCTA----------------GTGAAC |
| droSim1 | chr2L:20179920-20180047 + | CAT--------TCGAGAA---------------------------TTTGCACC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-TTAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGCTGACATCGAAAG-CCGA----------------GCGAAC |
| droSec1 | super_18:496867-496993 + | CAT--------TCGAGA----------------------------TTTGCACC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-TTAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGCTGACATCGAAAG-CCTA----------------GCGAAC |
| droYak2 | chr2R:6964793-6964921 + | TAT--------TCCAGAA---------------------------TTTCCTCC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-TTCCAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGATGACAACGAAGT-ACGA----------------GTAAAC |
| droEre2 | scaffold_4845:3836606-3836724 - | AT---------------------------------------------TTCCTC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-CCTAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGATGCCAACTAAAT-TCGA----------------GTGAAC |
| droAna3 | scaffold_12916:7117508-7117634 + | T----------TCGTGAA---------------------------TGATTATC--------TGCAAC-GCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATCT-TTTCAAGCATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATGACAACAAAGT-ACAA----------------GTGAAA |
| dp4 | chr4_group3:7699977-7700121 - | GAC--------GCGTGTG---------------------------TGCTTATG--------TACAACGGCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATTT-TATCAAGTATTTGGTCCCCTTCAACCAGCTGTAT-CAGTGGTTGATTCCAACAAAAA-ACAAAAGGCGCCAACAAATAATAAAG |
| droPer1 | super_1:9145698-9145865 - | TAT--------TTGTATGTGTAT--TT--GTATGTGACGCGTGTGTGCTTATG--------TACAACGGCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATTT-TATCAAGTATTTGGTCCCCTTCAACCAGCTGTAT-CAGTGGTTGATTCCAACAAAAA-ACAAAAGGCGCCAACAAATAATAAAG |
| droWil1 | scaffold_180708:2027542-2027651 + | -------------------------------------------------------------TACAAC-ACTGAATGTAGCTGGTTGACATCGGGTCAGATCTATTTATTTGAAGCATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATCCCAACAAAAA-GCAA-----------------TGGAA |
| droVir3 | scaffold_12963:13475783-13475914 - | CAA--------ACGAAGATATAT--ATTGCCAA--------TCGAATCACATT--------CACAAC-GCTGTGTGTAGTTGGTTGACATCGGGTCAGATCTATTT-CTGAAAGCATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGAAGCCAACAA------------------------------ |
| droMoj3 | scaffold_6500:24707638-24707772 + | CATGAAATAAATTGTGT------------------------TGTGTACTCAAACAAACATCCACAAC-GCTGTGTGTAGTTGGTTGACATCGGGTCAGATCTGTTT-T-TCGAGAATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATGACAACAAGT---------------------------- |
| droGri2 | scaffold_15126:6389834-6389991 + | CAA--------ACAAAACTATATACATGATTAA--------TTTGAATTTATT--------TACAAC-GCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATTT-ATTCGAGTATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATGTCAACAAGTGAACTTAAG-----AAC-AATAGTGAAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:20616602-20616729 + | CAT--------TCGAGAA---------------------------TTTCCACC--------TGCAAC-ACTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-TCAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGATGACAACGAAAG-CCTA----------------GTGAAC |
| droSim1 | chr2L:20179920-20180047 + | CAT--------TCGAGAA---------------------------TTTGCACC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-TTAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGCTGACATCGAAAG-CCGA----------------GCGAAC |
| droSec1 | super_18:496867-496993 + | CAT--------TCGAGA----------------------------TTTGCACC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-TTAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGCTGACATCGAAAG-CCTA----------------GCGAAC |
| droYak2 | chr2R:6964793-6964921 + | TAT--------TCCAGAA---------------------------TTTCCTCC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-TTCCAAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGATGACAACGAAGT-ACGA----------------GTAAAC |
| droEre2 | scaffold_4845:3836606-3836724 - | AT---------------------------------------------TTCCTC--------TGCAAC-GCTGTGTGTAGCTGGTTGACATCGGGTCAGATCTGTTT-T-CCTAGCATTTGGTCCCCTTCAACCAGCTGTAGCCAGTGGTTGATGCCAACTAAAT-TCGA----------------GTGAAC |
| droAna3 | scaffold_12916:7117508-7117634 + | T----------TCGTGAA---------------------------TGATTATC--------TGCAAC-GCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATCT-TTTCAAGCATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATGACAACAAAGT-ACAA----------------GTGAAA |
| dp4 | chr4_group3:7699977-7700121 - | GAC--------GCGTGTG---------------------------TGCTTATG--------TACAACGGCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATTT-TATCAAGTATTTGGTCCCCTTCAACCAGCTGTAT-CAGTGGTTGATTCCAACAAAAA-ACAAAAGGCGCCAACAAATAATAAAG |
| droPer1 | super_1:9145698-9145865 - | TAT--------TTGTATGTGTAT--TT--GTATGTGACGCGTGTGTGCTTATG--------TACAACGGCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATTT-TATCAAGTATTTGGTCCCCTTCAACCAGCTGTAT-CAGTGGTTGATTCCAACAAAAA-ACAAAAGGCGCCAACAAATAATAAAG |
| droWil1 | scaffold_180708:2027542-2027651 + | -------------------------------------------------------------TACAAC-ACTGAATGTAGCTGGTTGACATCGGGTCAGATCTATTTATTTGAAGCATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATCCCAACAAAAA-GCAA-----------------TGGAA |
| droVir3 | scaffold_12963:13475783-13475914 - | CAA--------ACGAAGATATAT--ATTGCCAA--------TCGAATCACATT--------CACAAC-GCTGTGTGTAGTTGGTTGACATCGGGTCAGATCTATTT-CTGAAAGCATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGAAGCCAACAA------------------------------ |
| droMoj3 | scaffold_6500:24707638-24707772 + | CATGAAATAAATTGTGT------------------------TGTGTACTCAAACAAACATCCACAAC-GCTGTGTGTAGTTGGTTGACATCGGGTCAGATCTGTTT-T-TCGAGAATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATGACAACAAGT---------------------------- |
| droGri2 | scaffold_15126:6389834-6389991 + | CAA--------ACAAAACTATATACATGATTAA--------TTTGAATTTATT--------TACAAC-GCTGTATGTAGCTGGTTGACATCGGGTCAGATCTATTT-ATTCGAGTATTTGGTCCCCTTCAACCAGCTGTAG-CAGTGGTTGATGTCAACAAGTGAACTTAAG-----AAC-AATAGTGAAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
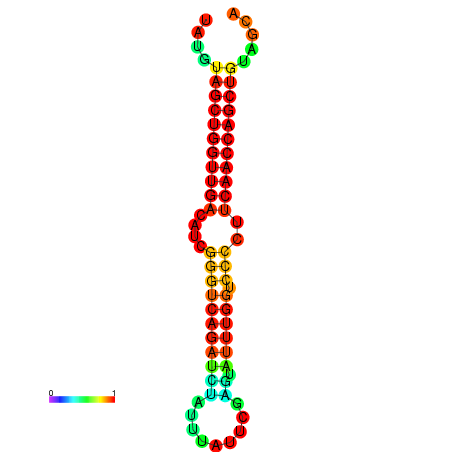
| dG=-27.2, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.6, p-value=0.009901 |
|---|---|---|
 |
 |
 |
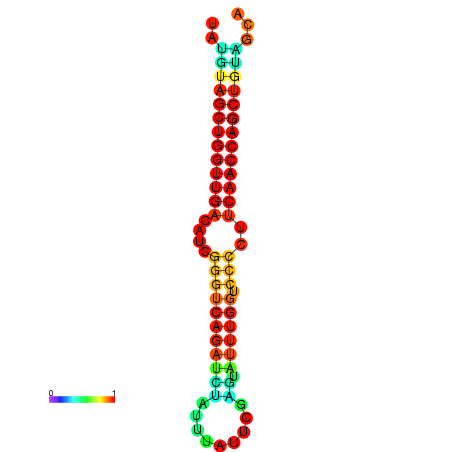
| dG=-27.2, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.6, p-value=0.009901 |
|---|---|---|
 |
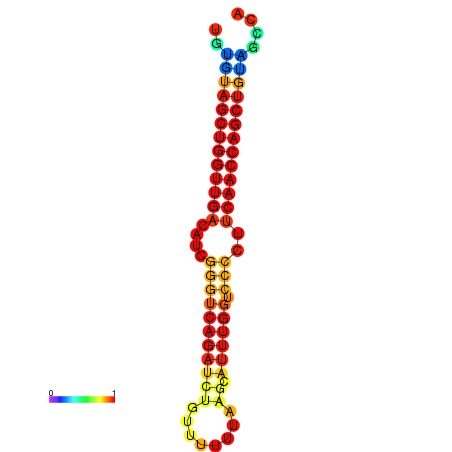 |
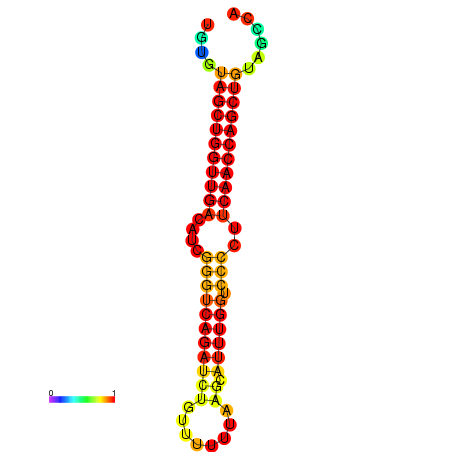 |
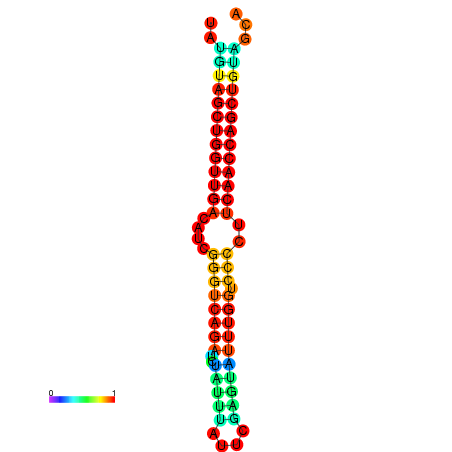
| dG=-27.2, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.6, p-value=0.009901 |
|---|---|---|
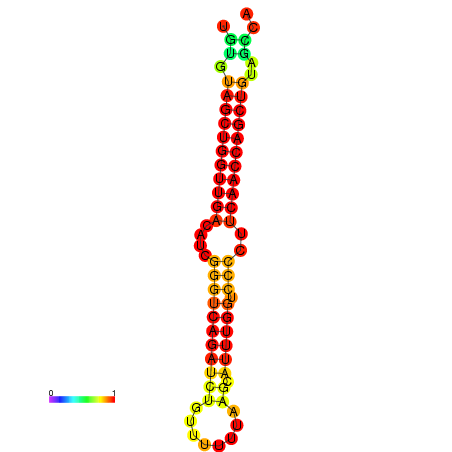 |
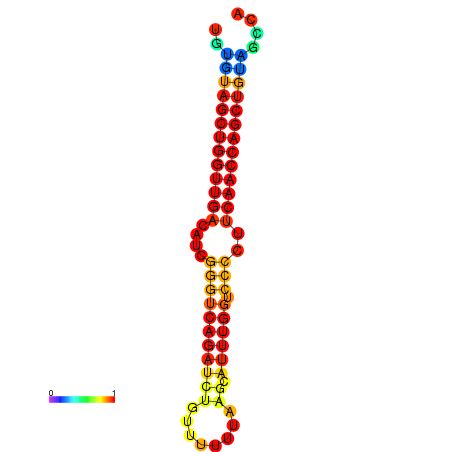 |
 |
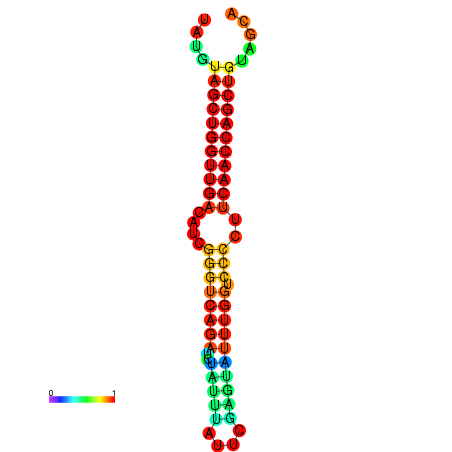
| dG=-26.3, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|---|
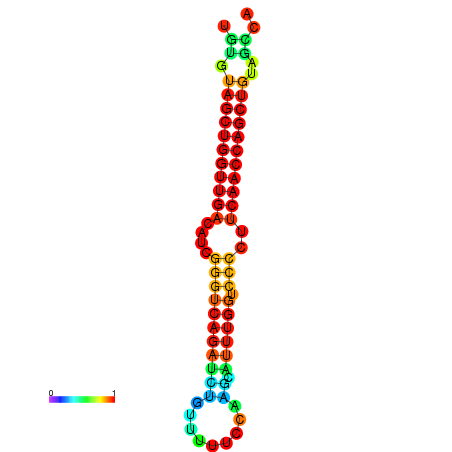 |
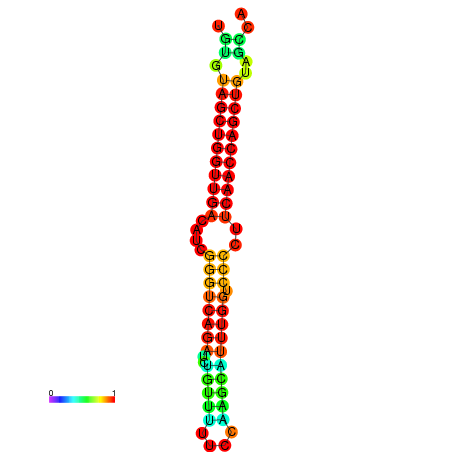 |
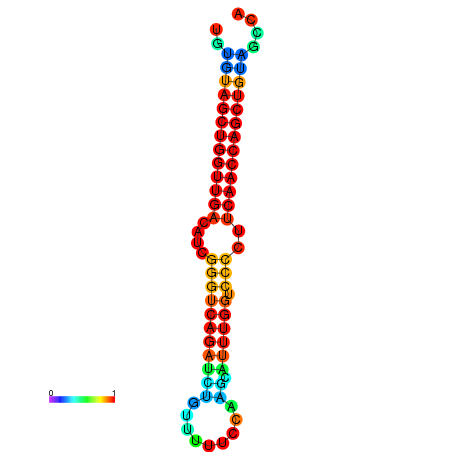 |
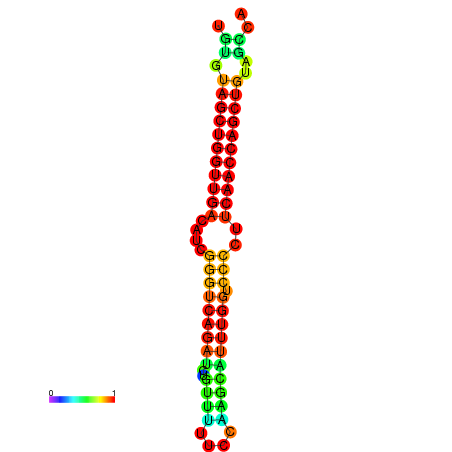 |
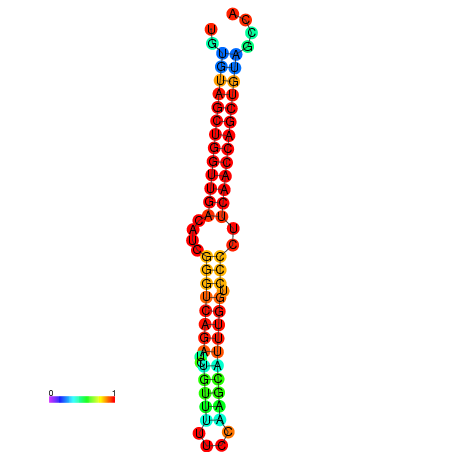 |
| dG=-27.1, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|
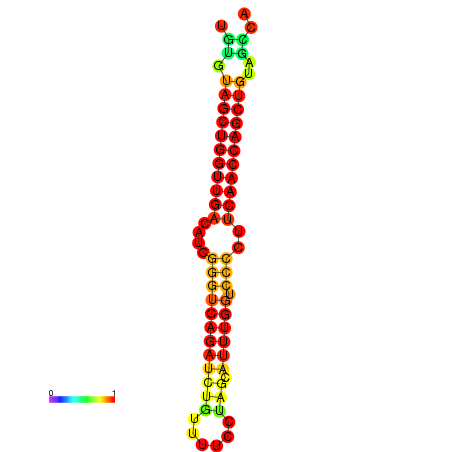 |
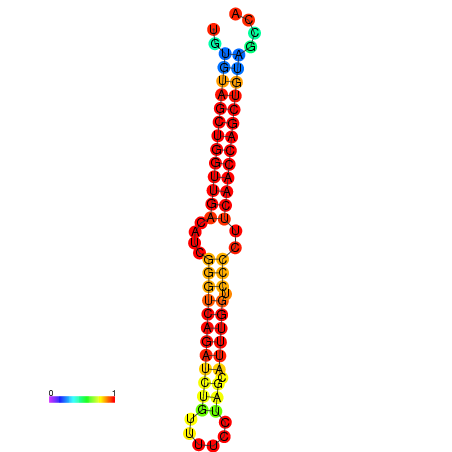 |
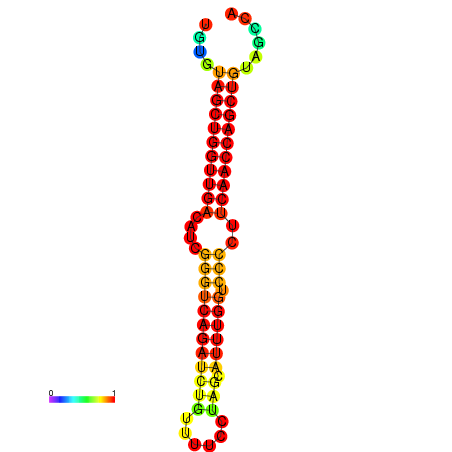 |
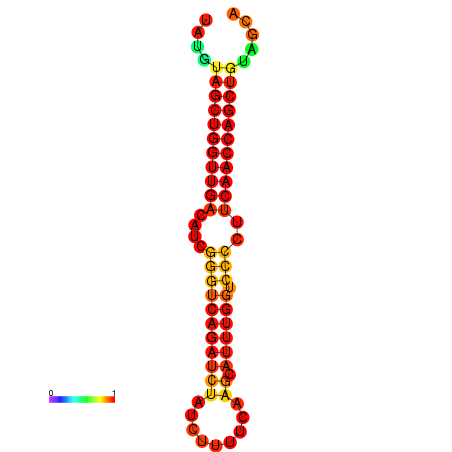
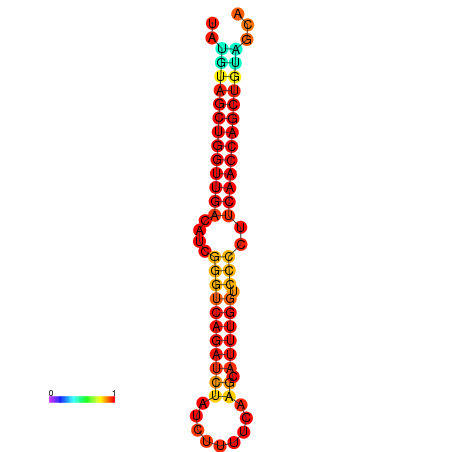
| dG=-24.7, p-value=0.009901 | dG=-24.7, p-value=0.009901 |
|---|---|
 |
 |
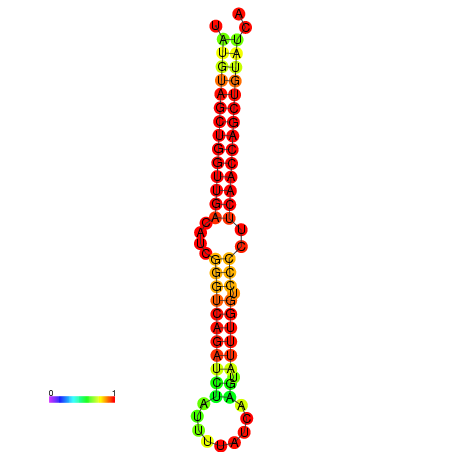
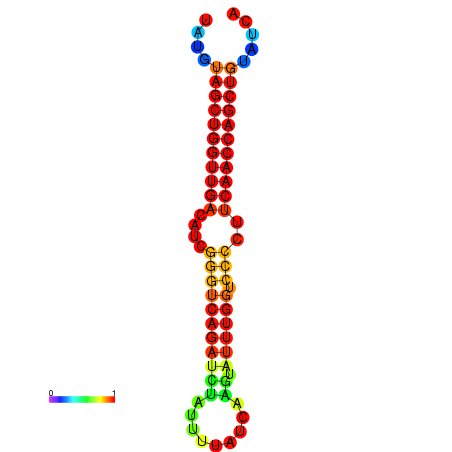
| dG=-25.5, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|
 |
 |
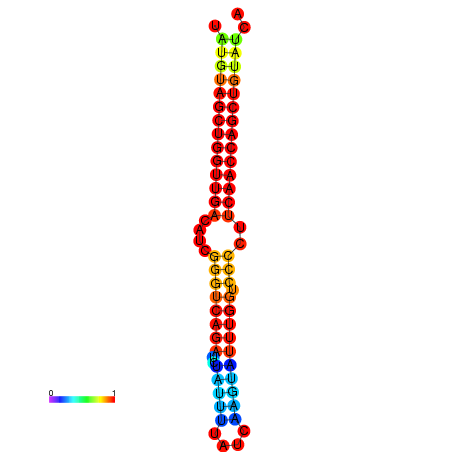 |
| dG=-25.5, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|
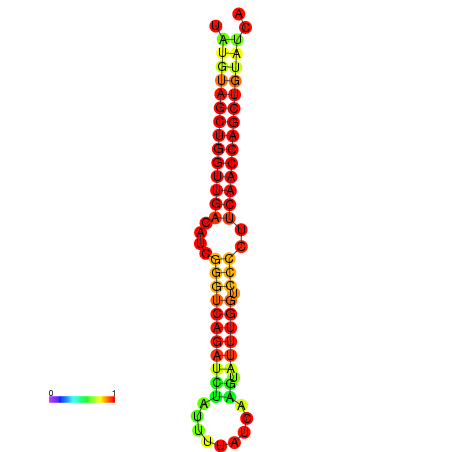 |
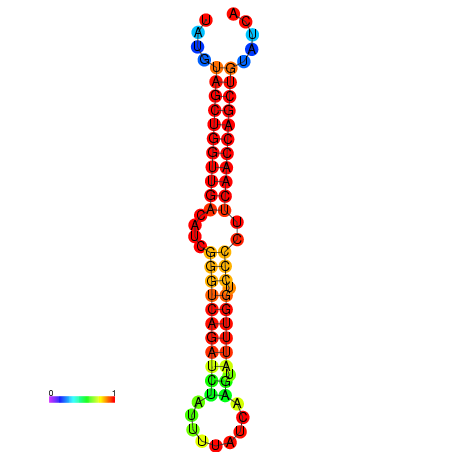 |
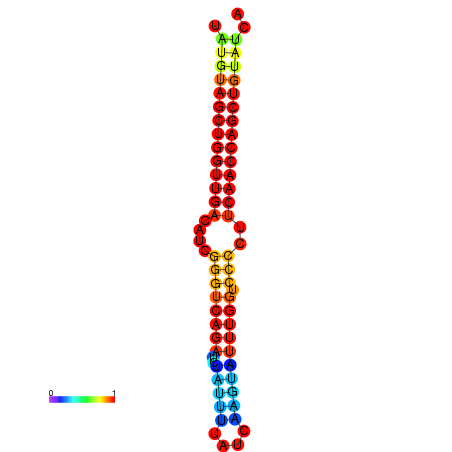 |
| dG=-24.6, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
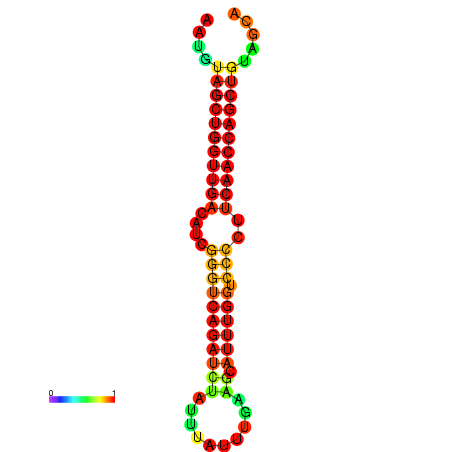 |
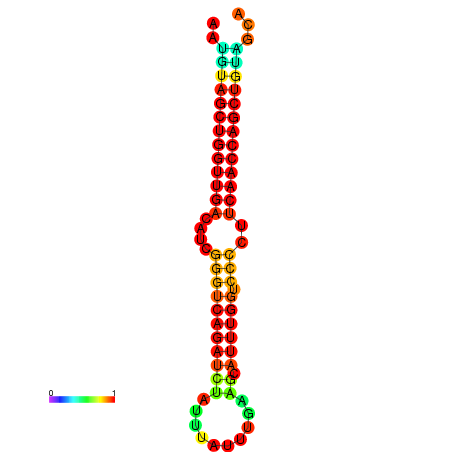 |
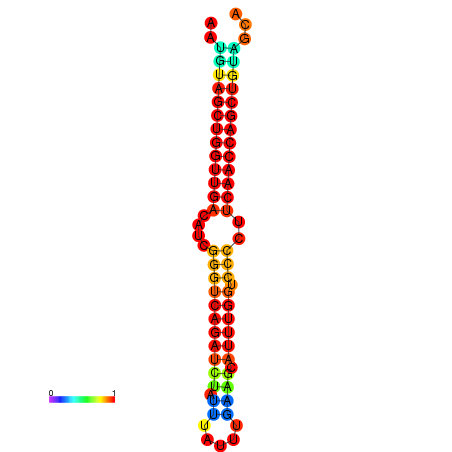 |
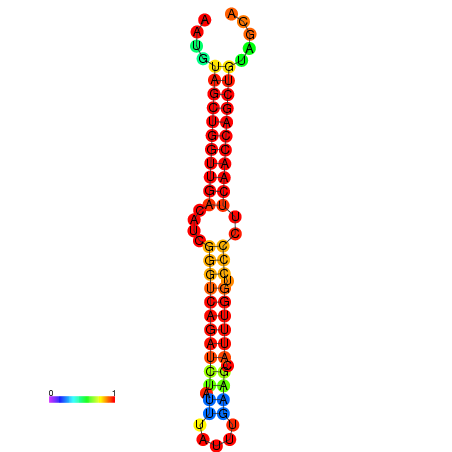 |
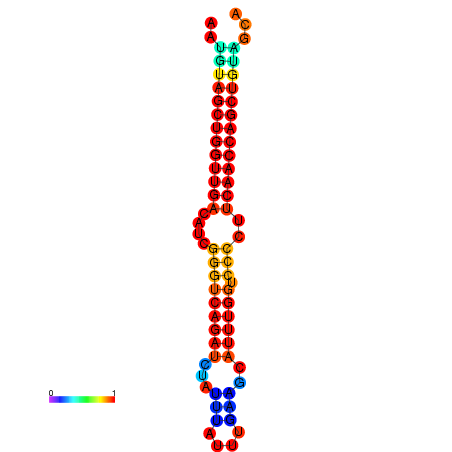 |
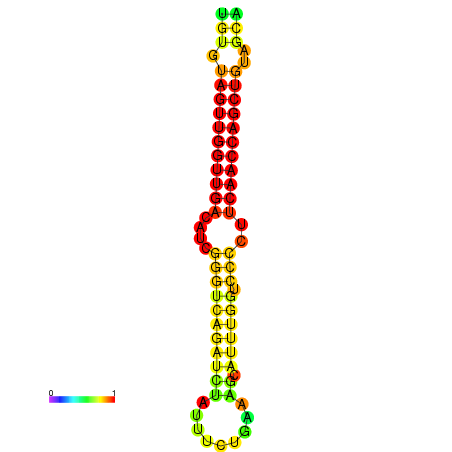
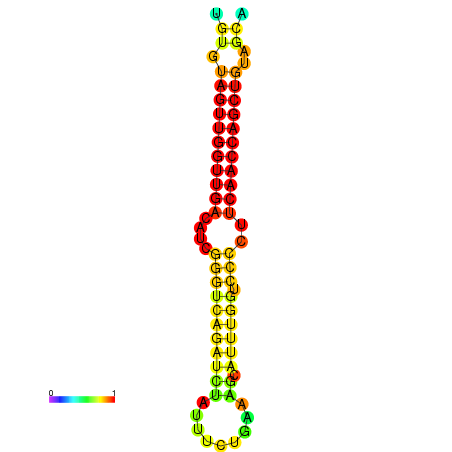
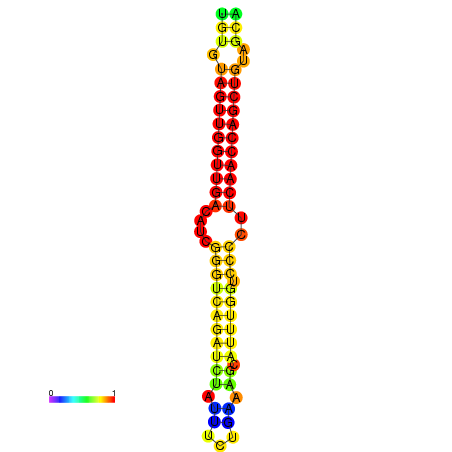
| dG=-23.5, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.9, p-value=0.009901 | dG=-22.7, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
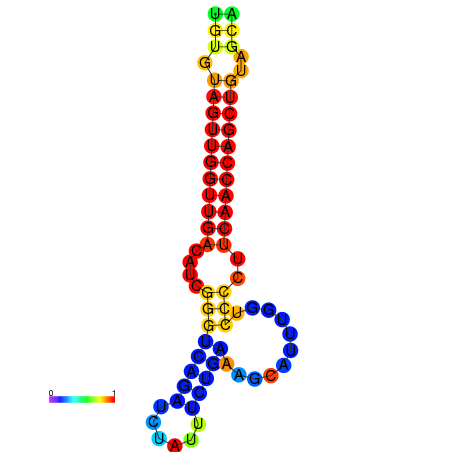 |
| dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|
 |
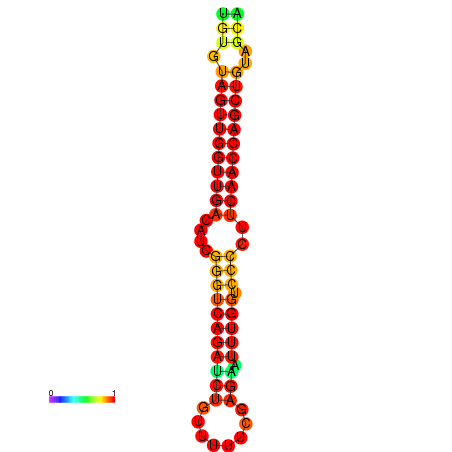 |
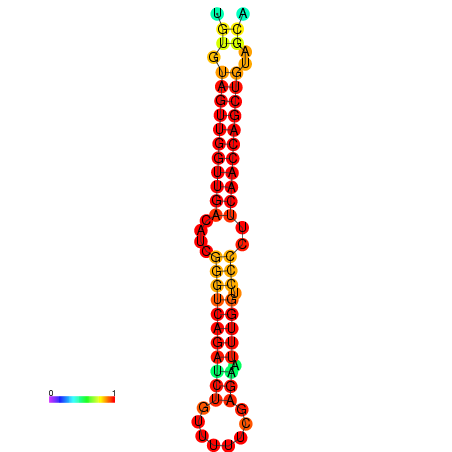 |
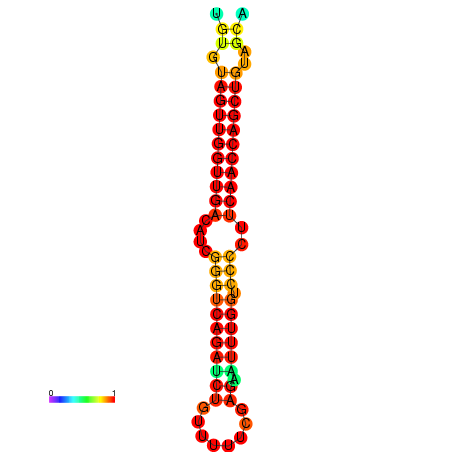 |
| dG=-24.7, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:50 PM