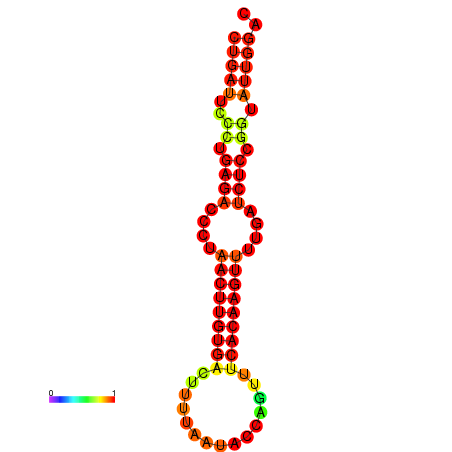
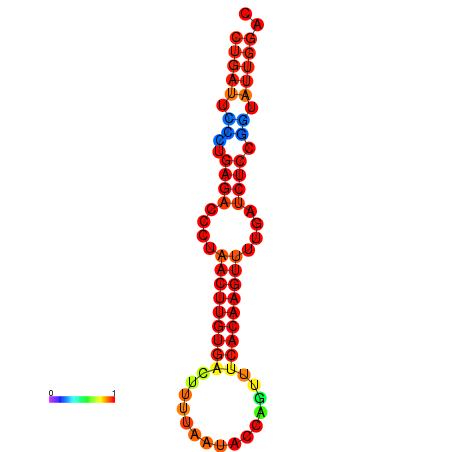
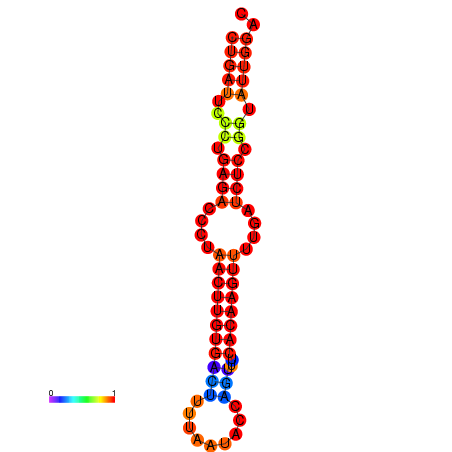
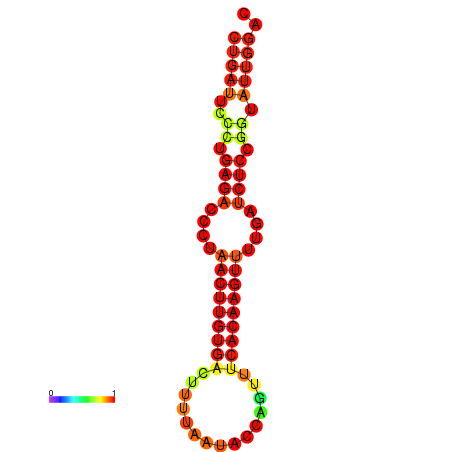
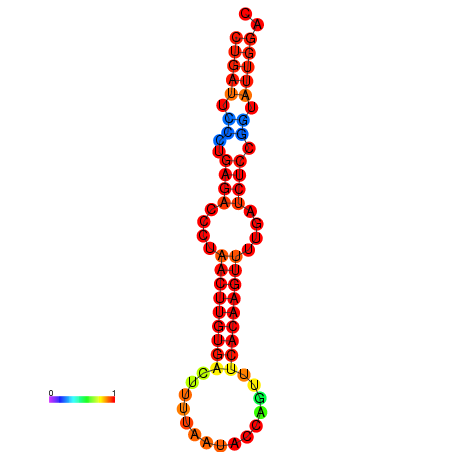
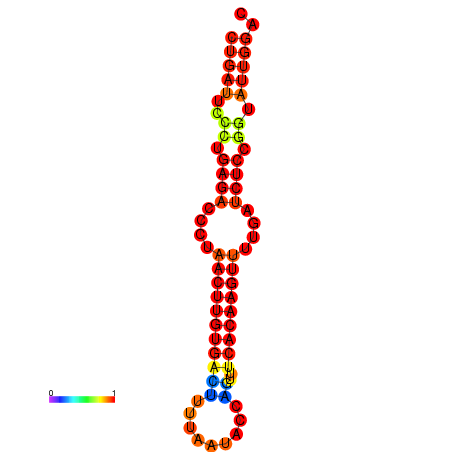
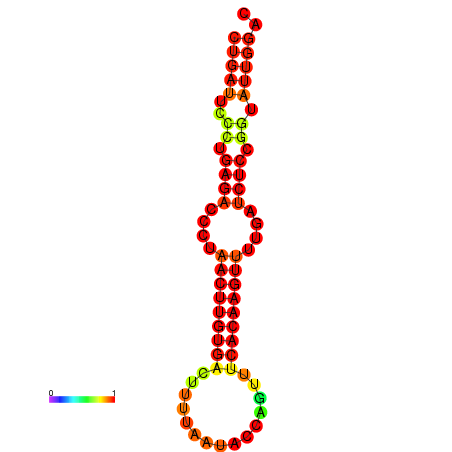
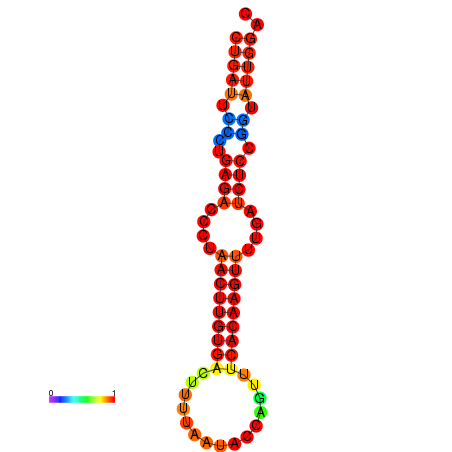
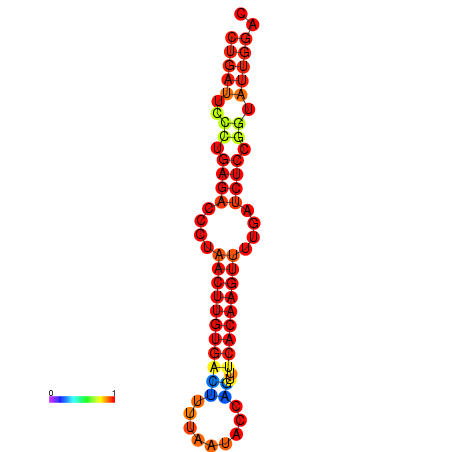
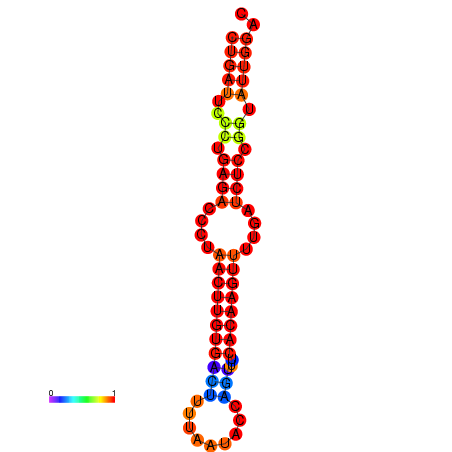
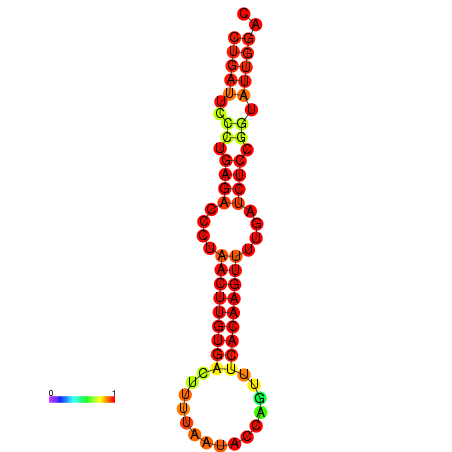
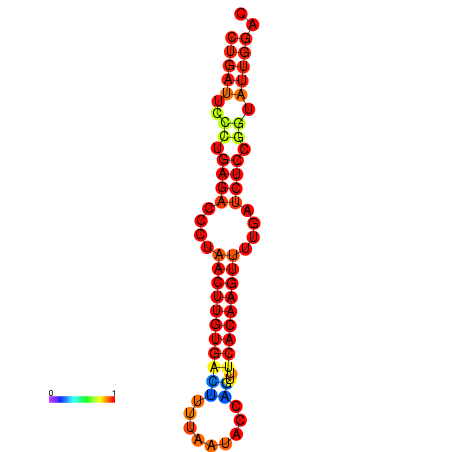
| AGTATTGGTATGAGTGACATGTGCAAATGTTTGTATGGCTGATTCCCTGAGACCCTAACTTGTGACTTTTAATACCAGTTTCACAAGTTTTGATCTCCGGTATTGGACGCAAACTTGCTGATGTTATACAAAAAAAAAAGC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ...........................................TCCCTGAGACCCTAACTTGTGA............................................................................ | 22 | 1 | 7563.00 | 7563 | 13 | 7550 |
| ...........................................TCCCTGAGACCCTAACTTGTG............................................................................. | 21 | 1 | 2246.00 | 2246 | 0 | 2246 |
| ...........................................TCCCTGAGACCCTAACTTGT.............................................................................. | 20 | 1 | 1091.00 | 1091 | 0 | 1091 |
| .............................................CCTGAGACCCTAACTTGTGA............................................................................ | 20 | 1 | 44.00 | 44 | 0 | 44 |
| ..................................................................................ACAAGTTTTGATCTCCGGTAT...................................... | 21 | 1 | 30.00 | 30 | 0 | 30 |
| ...........................................TCCCTGAGACCCTAACTTG............................................................................... | 19 | 1 | 28.00 | 28 | 0 | 28 |
| .............................................CCTGAGACCCTAACTTGTG............................................................................. | 19 | 1 | 14.00 | 14 | 0 | 14 |
| ...........................................TCCCTGAGACCCTAACTTGTGACT.......................................................................... | 24 | 1 | 9.00 | 9 | 0 | 9 |
| ..............................................CTGAGACCCTAACTTGTGA............................................................................ | 19 | 1 | 8.00 | 8 | 0 | 8 |
| .............................................CCTGAGACCCTAACTTGT.............................................................................. | 18 | 1 | 6.00 | 6 | 0 | 6 |
| ............................................CCCTGAGACCCTAACTTGTGA............................................................................ | 21 | 1 | 4.00 | 4 | 0 | 4 |
| ...........................................TCCCTGAGACCCTAACTTGTGAC........................................................................... | 23 | 1 | 3.00 | 3 | 0 | 3 |
| ...........................................TCCCTGAGACCCTAACTT................................................................................ | 18 | 1 | 2.00 | 2 | 0 | 2 |
| ..................................................................................ACAAGTTTTGATCTCCGGT........................................ | 19 | 1 | 2.00 | 2 | 0 | 2 |
| ..............................................CTGAGACCCTAACTTGTG............................................................................. | 18 | 1 | 2.00 | 2 | 0 | 2 |
| ................................................................................TCACAAGTTTTGATCTCCGGTAT...................................... | 23 | 1 | 2.00 | 2 | 0 | 2 |
| ...........................................TCCCTGAGACCCTAACTTGTGACTT......................................................................... | 25 | 1 | 2.00 | 2 | 0 | 2 |
| ................................................................................TCACAAGTTTTGATCTCCGGTA....................................... | 22 | 1 | 1.00 | 1 | 0 | 1 |
| ..................................................................................ACAAGTTTTGATCTCCGGTA....................................... | 20 | 1 | 1.00 | 1 | 0 | 1 |
| ..............................................CTGAGACCCTAACTTGTGACT.......................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 |
| ..................................................................................ACAAGTTTTGATCTCCGGTATT..................................... | 22 | 1 | 1.00 | 1 | 0 | 1 |