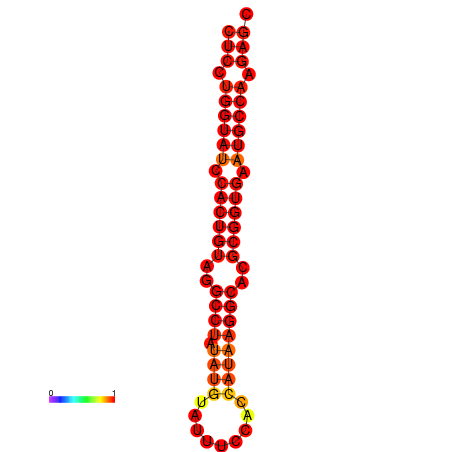
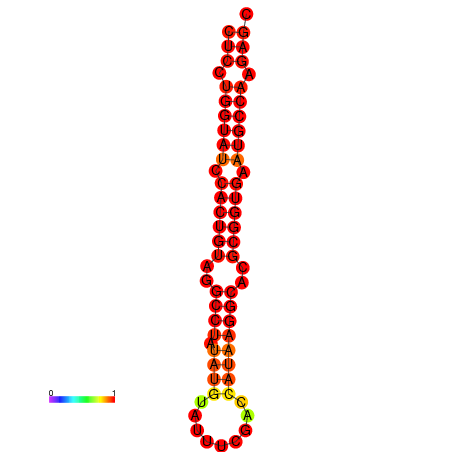
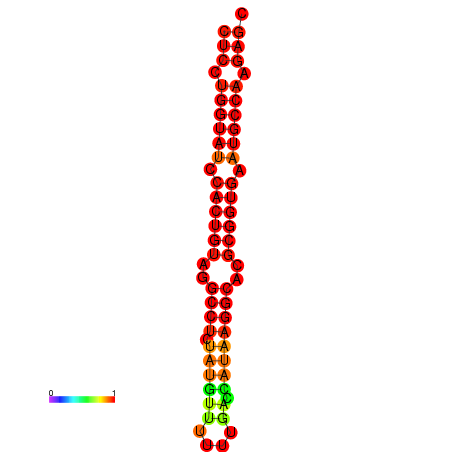
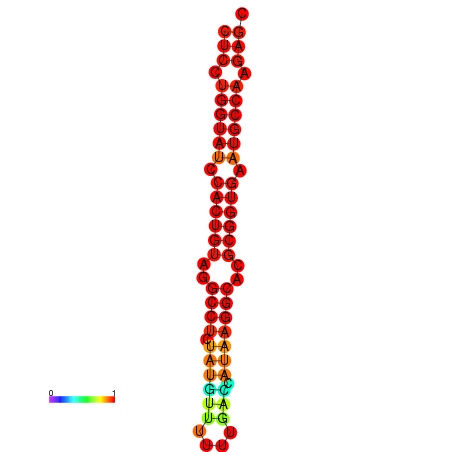
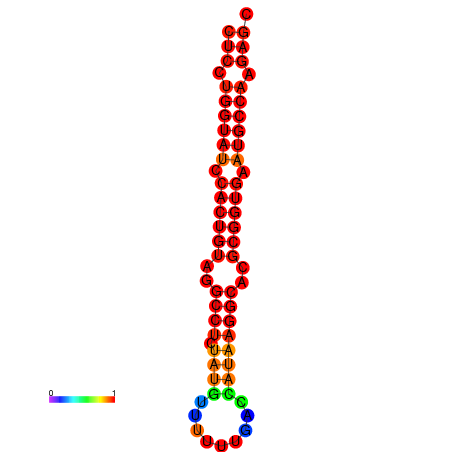
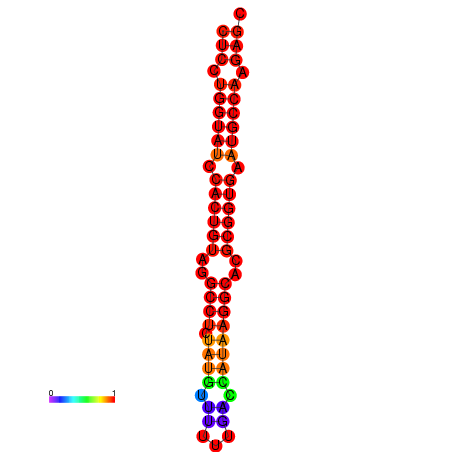
| TCTCAGTGC---TCGTCGTTTGGTACGTTTTTCTCCTGGTATCCACTGTAGGCCTATATGTATTTCGACCATAAGGCACGCGGTGAATGCCAAGAGCGGACGAAACTCTACTA-----TCAAAAACAGCA | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| .......................................................................TAAGGCACGCGGTGAATGCCA...................................... | 21 | 1 | 37470.00 | 37470 | 3038 | 34432 |
| .......................................................................TAAGGCACGCGGTGAATGCCAAG.................................... | 23 | 1 | 10075.00 | 10075 | 4518 | 5557 |
| .......................................................................TAAGGCACGCGGTGAATGCCAA..................................... | 22 | 1 | 5869.00 | 5869 | 2270 | 3599 |
| .......................................................................TAAGGCACGCGGTGAATGCC....................................... | 20 | 1 | 5677.00 | 5677 | 207 | 5470 |
| ....................................TGGTATCCACTGTAGGCCTATATG...................................................................... | 24 | 1 | 466.00 | 466 | 346 | 120 |
| .......................................................................TAAGGCACGCGGTGAATGC........................................ | 19 | 1 | 256.00 | 256 | 4 | 252 |
| .....................................GGTATCCACTGTAGGCCTATATG...................................................................... | 23 | 1 | 208.00 | 208 | 194 | 14 |
| .......................................................................TAAGGCACGCGGTGAATGCCAAGA................................... | 24 | 1 | 195.00 | 195 | 24 | 171 |
| ........................................................................AAGGCACGCGGTGAATGCCA...................................... | 20 | 1 | 118.00 | 118 | 5 | 113 |
| ........................................................................AAGGCACGCGGTGAATGCCAAG.................................... | 22 | 1 | 96.00 | 96 | 45 | 51 |
| .......................................................................TAAGGCACGCGGTGAATG......................................... | 18 | 1 | 41.00 | 41 | 3 | 38 |
| ....................................TGGTATCCACTGTAGGCCTATA........................................................................ | 22 | 1 | 27.00 | 27 | 26 | 1 |
| ....................................TGGTATCCACTGTAGGCCTA.......................................................................... | 20 | 1 | 25.00 | 25 | 1 | 24 |
| ....................................TGGTATCCACTGTAGGCCTATAT....................................................................... | 23 | 1 | 25.00 | 25 | 18 | 7 |
| ........................................................................AAGGCACGCGGTGAATGCCAA..................................... | 21 | 1 | 18.00 | 18 | 5 | 13 |
| .....................................GGTATCCACTGTAGGCCTATA........................................................................ | 21 | 1 | 15.00 | 15 | 15 | 0 |
| ........................................................................AAGGCACGCGGTGAATGCC....................................... | 19 | 1 | 15.00 | 15 | 0 | 15 |
| .........................................................................AGGCACGCGGTGAATGCCAAG.................................... | 21 | 1 | 13.00 | 13 | 7 | 6 |
| .........................................................................AGGCACGCGGTGAATGCCA...................................... | 19 | 1 | 10.00 | 10 | 2 | 8 |
| ....................................TGGTATCCACTGTAGGCCTAT......................................................................... | 21 | 1 | 9.00 | 9 | 5 | 4 |
| ....................................TGGTATCCACTGTAGGCCT........................................................................... | 19 | 1 | 8.00 | 8 | 0 | 8 |
| .....................................GGTATCCACTGTAGGCCTATAT....................................................................... | 22 | 1 | 7.00 | 7 | 6 | 1 |
| .........................................................................AGGCACGCGGTGAATGCCAA..................................... | 20 | 1 | 6.00 | 6 | 2 | 4 |