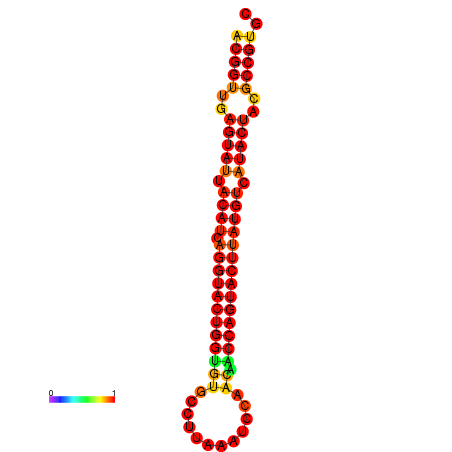
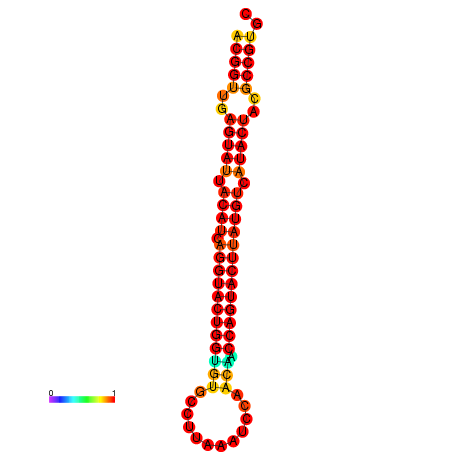

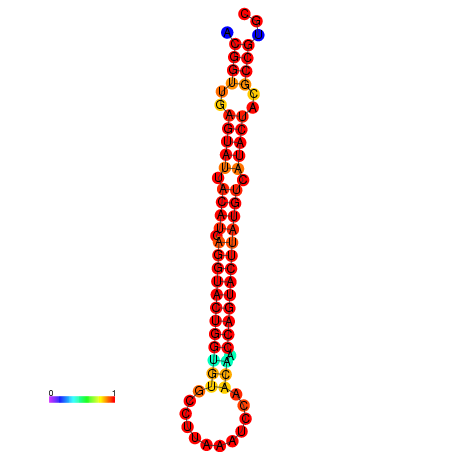
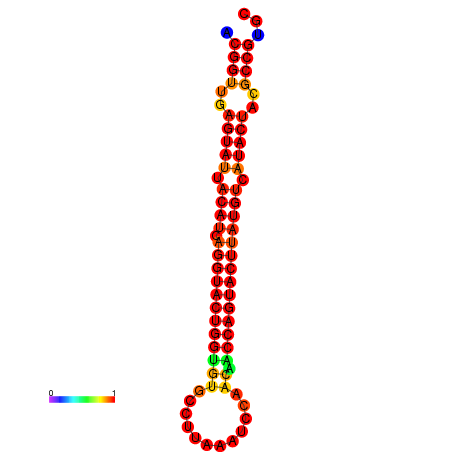
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:15403491-15403594 + | AA-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droSim1 | chrX_random:4024315-4024418 + | AT-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTGAAATCCAACAACCAGTACTTGTGTCATACTACGCCGTGCACGGATCGCACTAA |
| droSec1 | super_73:137914-138017 + | AT-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTGAAATCCAACAACCAGTACTTGTGTCATACTACGCCGTGCACGGATCGCACTAA |
| droYak2 | gnl|ti|371645083:614-718 + | AA-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droEre2 | scaffold_4690:11630938-11631041 - | AA-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAAACCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droAna3 | scaffold_12929:2898571-2898667 - | -------------------------------GCGTCTGTACGGCTGAGTATTACATCAGGTACTGGTGTGCCTTTAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACCGATCGCACTAA |
| dp4 | chrXL_group1a:977995-978098 + | AT-----------------CCAG------CAGCGTCTGTACGGCTGAGTATTACATCAGGTACTGGTGTGCTTTAAATCCAACAACCAGTACTTATGTTATACTACGCCGTGCACTGATCGCACTAA |
| droPer1 | super_26:1092636-1092739 + | AT-----------------CCAG------CAGCGTCTGTACGGCTGAGTATTACATCAGGTACTGGTGTGCTTTAAATCCAACAACCAGTACTTATGTTATACTACGCCGTGCACTGATCGCACTAA |
| droWil1 | scaffold_180702:1592742-1592841 - | ----------------------G------CAGCGCCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTTAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droVir3 | scaffold_12970:2049296-2049416 + | AACAGCCGCATCAGCATCCGCAT------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCATTTAACTAACAACCAGTACTTATGTCATACTACGCCGTGCAGTGGGCGCACTAA |
| droMoj3 | scaffold_6359:3259767-3259875 + | AG------------CATCAGCAT------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCAATTAACTAACAACCAGTACTTATGTCATACTACGCCGTGCAGTGGGCGCACTAA |
| droGri2 | scaffold_14853:6951707-6951816 - | AA-----------------GCAGCATCACAAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCATTTAACCAACAACCAGTACTTATGTCATACTACGCCGTGCAGTGGGCGCACTAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:15403491-15403594 + | AA-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droSim1 | chrX_random:4024315-4024418 + | AT-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTGAAATCCAACAACCAGTACTTGTGTCATACTACGCCGTGCACGGATCGCACTAA |
| droSec1 | super_73:137914-138017 + | AT-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTGAAATCCAACAACCAGTACTTGTGTCATACTACGCCGTGCACGGATCGCACTAA |
| droYak2 | gnl|ti|371645083:614-718 + | AA-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droEre2 | scaffold_4690:11630938-11631041 - | AA-----------------GGAG------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAAACCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droAna3 | scaffold_12929:2898571-2898667 - | -------------------------------GCGTCTGTACGGCTGAGTATTACATCAGGTACTGGTGTGCCTTTAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACCGATCGCACTAA |
| dp4 | chrXL_group1a:977995-978098 + | AT-----------------CCAG------CAGCGTCTGTACGGCTGAGTATTACATCAGGTACTGGTGTGCTTTAAATCCAACAACCAGTACTTATGTTATACTACGCCGTGCACTGATCGCACTAA |
| droPer1 | super_26:1092636-1092739 + | AT-----------------CCAG------CAGCGTCTGTACGGCTGAGTATTACATCAGGTACTGGTGTGCTTTAAATCCAACAACCAGTACTTATGTTATACTACGCCGTGCACTGATCGCACTAA |
| droWil1 | scaffold_180702:1592742-1592841 - | ----------------------G------CAGCGCCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTTAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA |
| droVir3 | scaffold_12970:2049296-2049416 + | AACAGCCGCATCAGCATCCGCAT------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCATTTAACTAACAACCAGTACTTATGTCATACTACGCCGTGCAGTGGGCGCACTAA |
| droMoj3 | scaffold_6359:3259767-3259875 + | AG------------CATCAGCAT------CAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCAATTAACTAACAACCAGTACTTATGTCATACTACGCCGTGCAGTGGGCGCACTAA |
| droGri2 | scaffold_14853:6951707-6951816 - | AA-----------------GCAGCATCACAAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCATTTAACCAACAACCAGTACTTATGTCATACTACGCCGTGCAGTGGGCGCACTAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
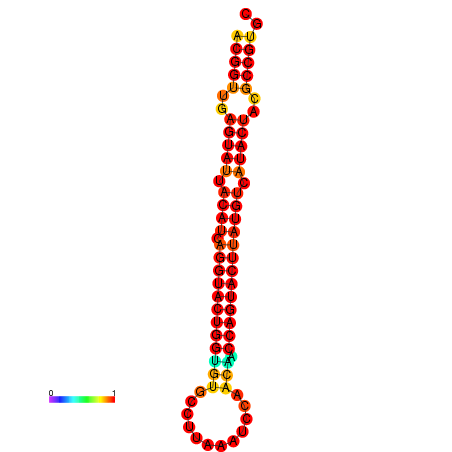
| dG=-31.3, p-value=0.009901 | dG=-31.3, p-value=0.009901 | dG=-30.4, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
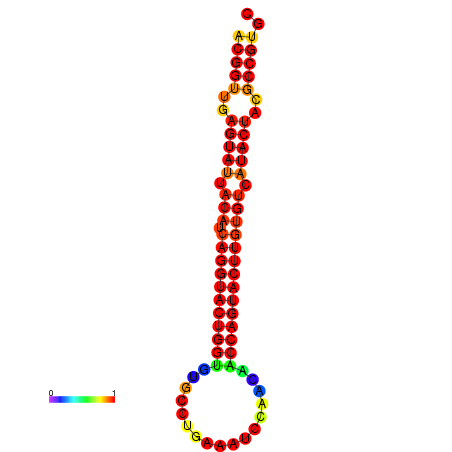
| dG=-31.3, p-value=0.009901 | dG=-31.3, p-value=0.009901 | dG=-30.4, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
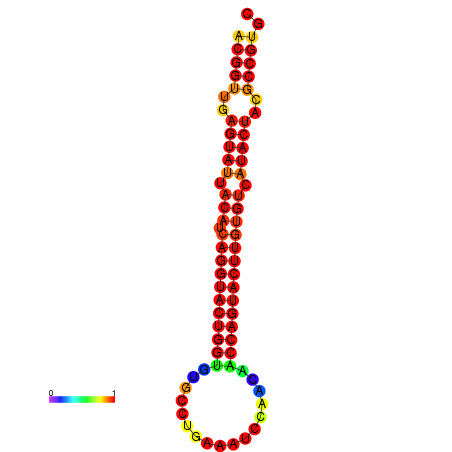
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
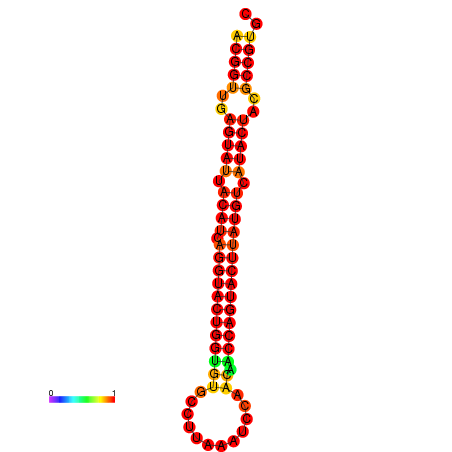 |
 |
 |
 |
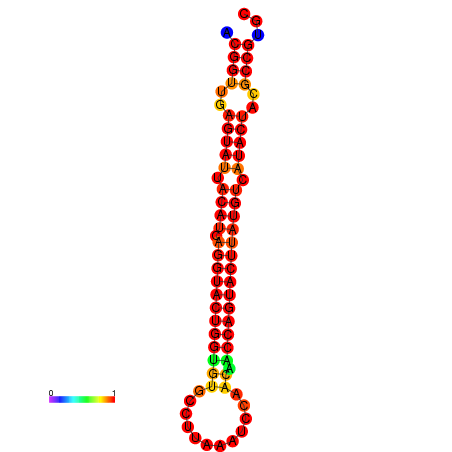 |
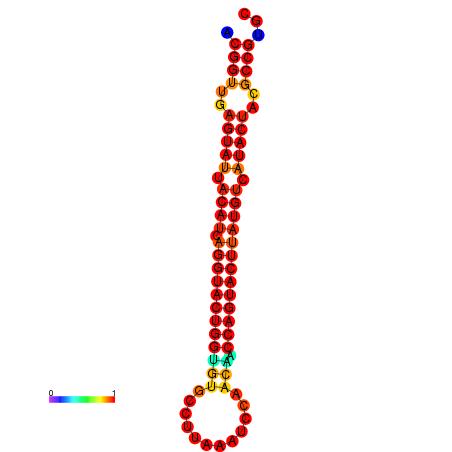
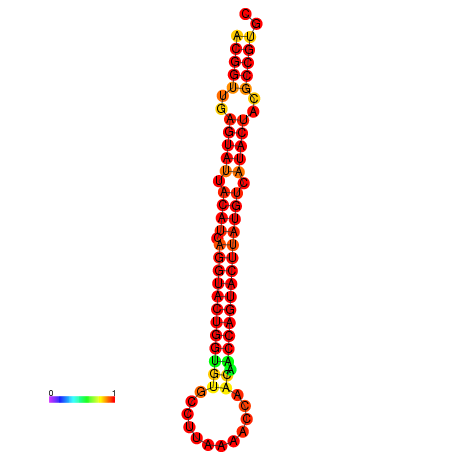
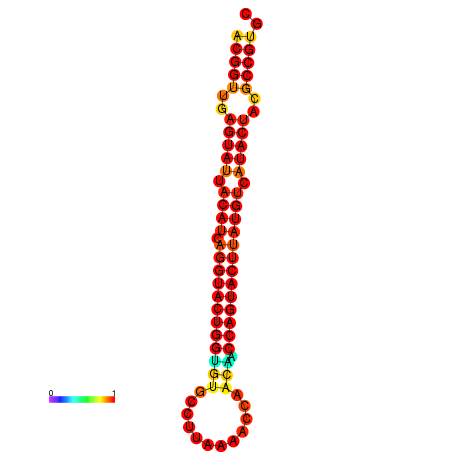
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
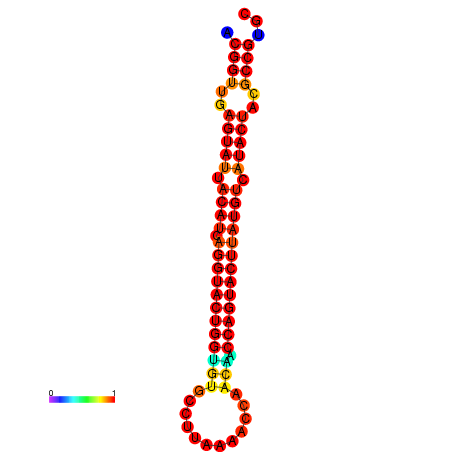 |
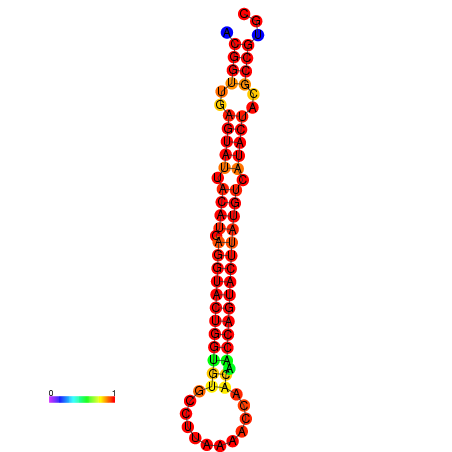 |
| dG=-31.0, p-value=0.009901 | dG=-31.0, p-value=0.009901 | dG=-30.1, p-value=0.009901 | dG=-30.0, p-value=0.009901 | dG=-30.0, p-value=0.009901 |
|---|---|---|---|---|
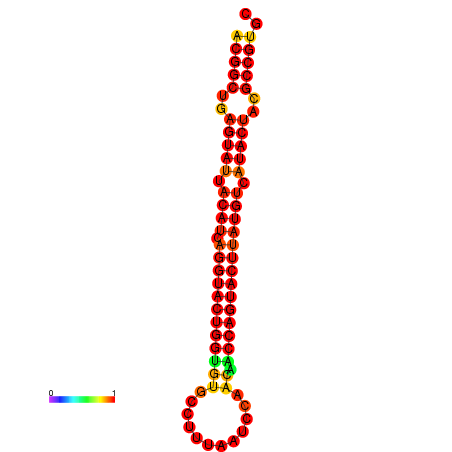 |
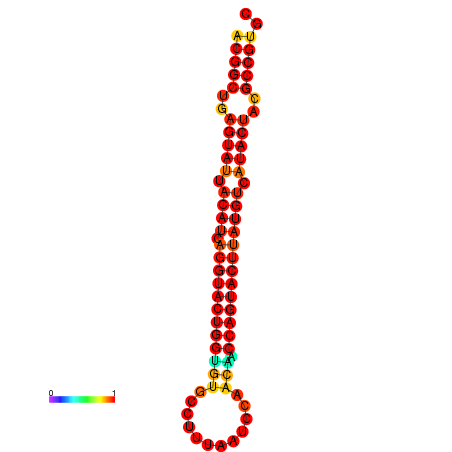 |
 |
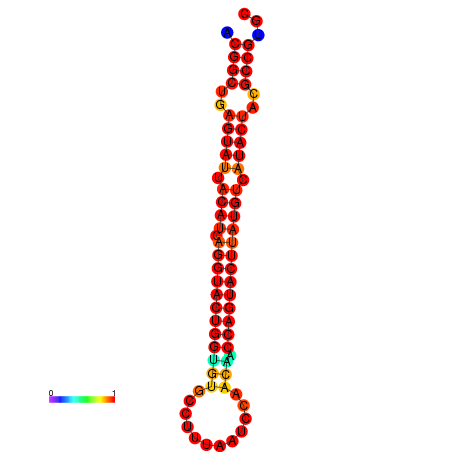 |
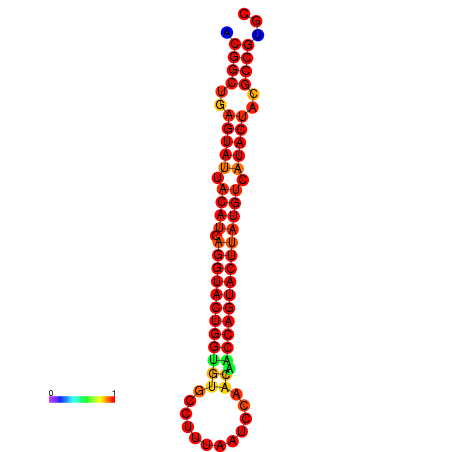 |
| dG=-31.2, p-value=0.009901 | dG=-31.2, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
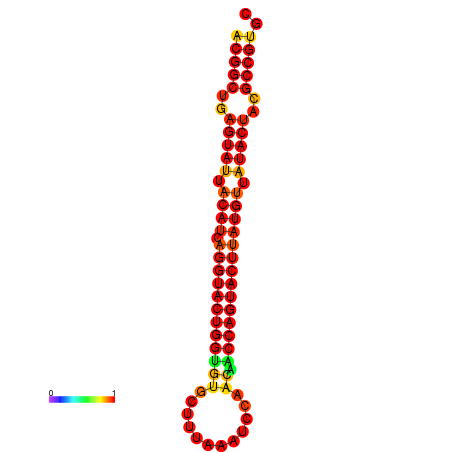 |
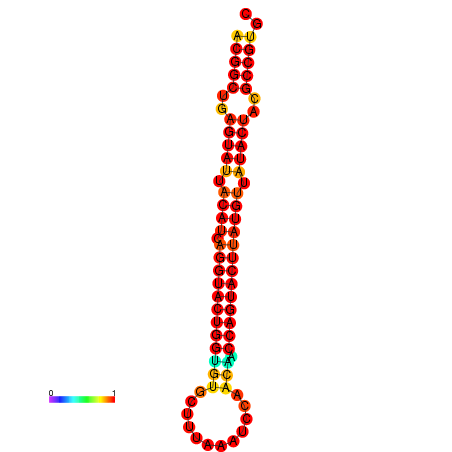 |
 |
 |
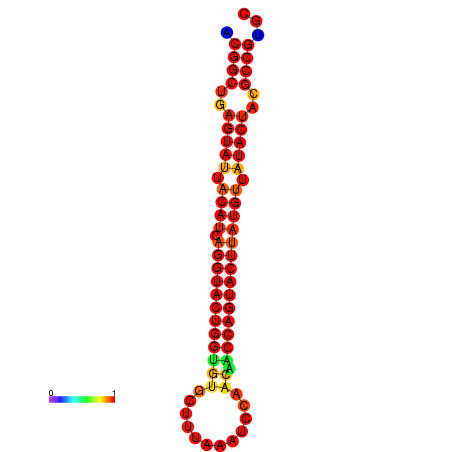 |
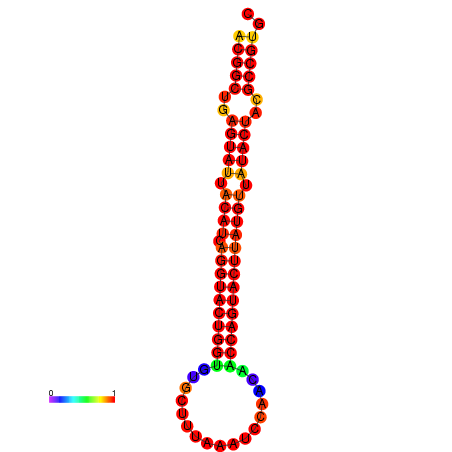
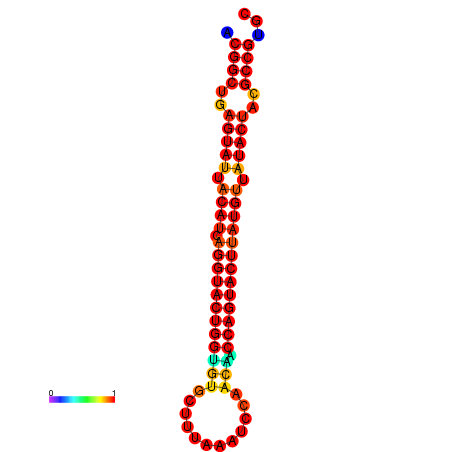
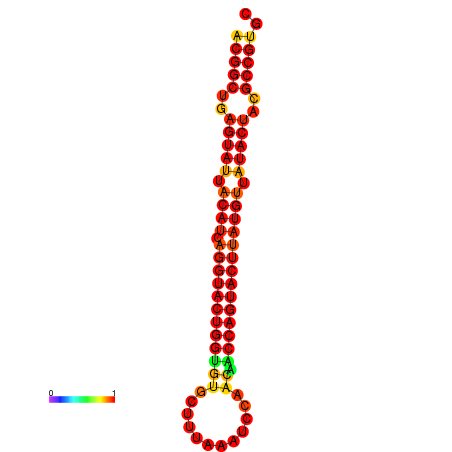
| dG=-31.2, p-value=0.009901 | dG=-31.2, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
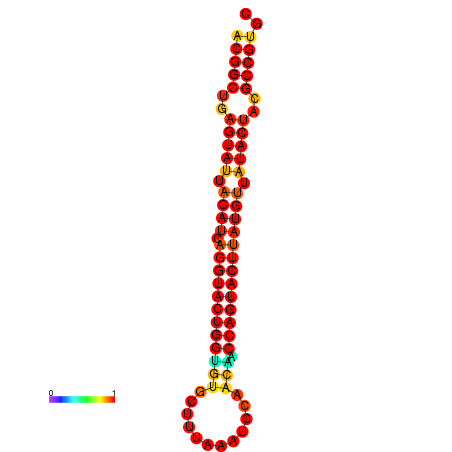
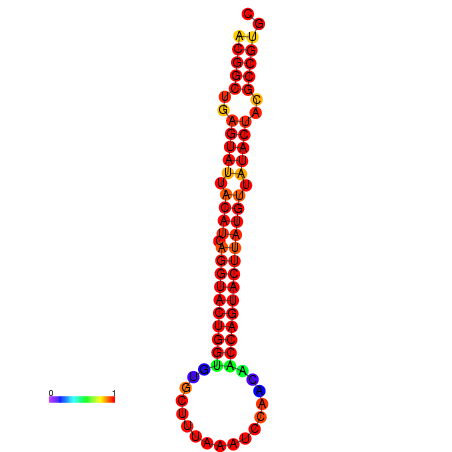
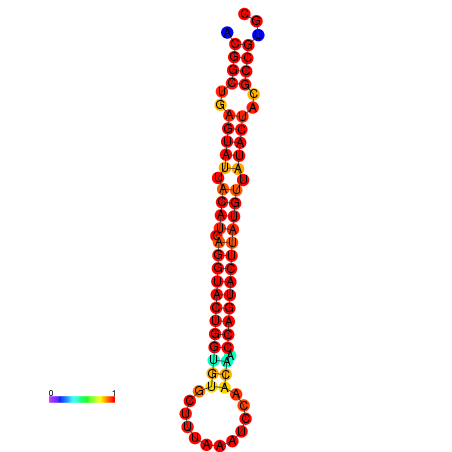
 |
 |
 |
 |
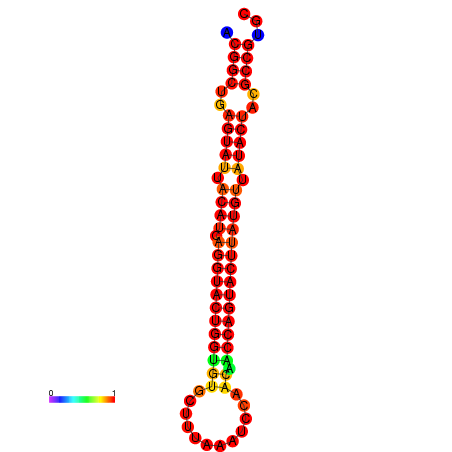 |
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
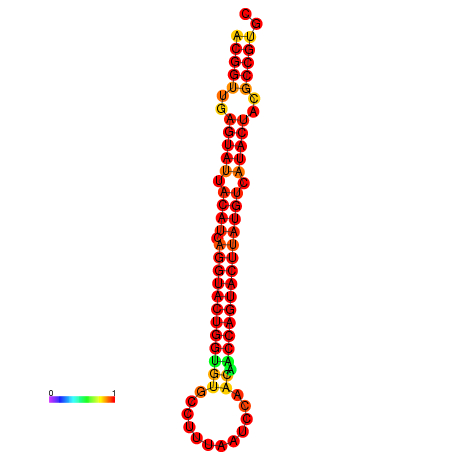 |
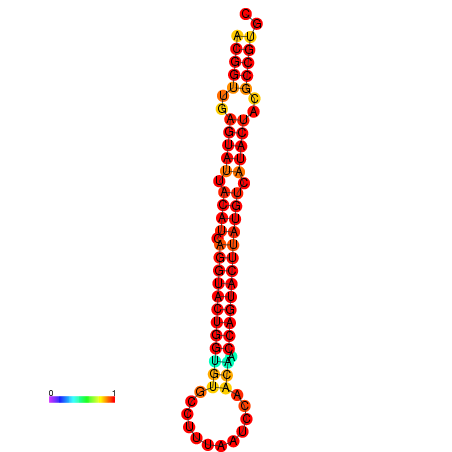 |
 |
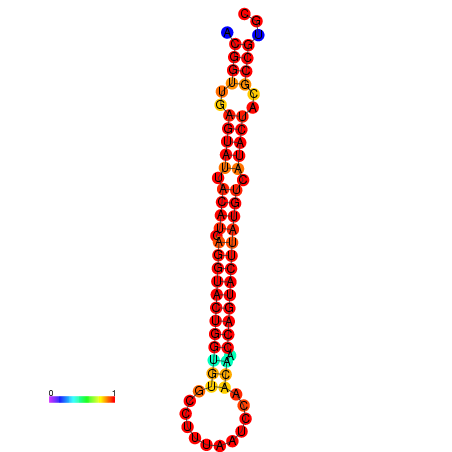 |
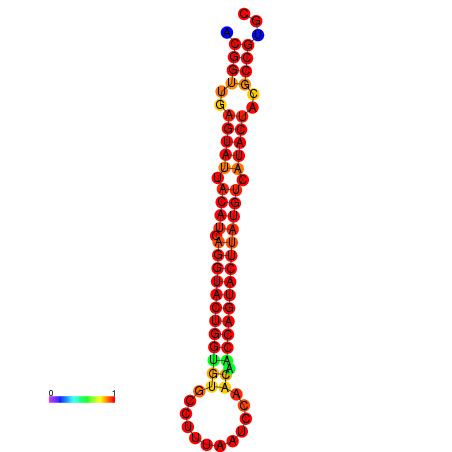 |
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
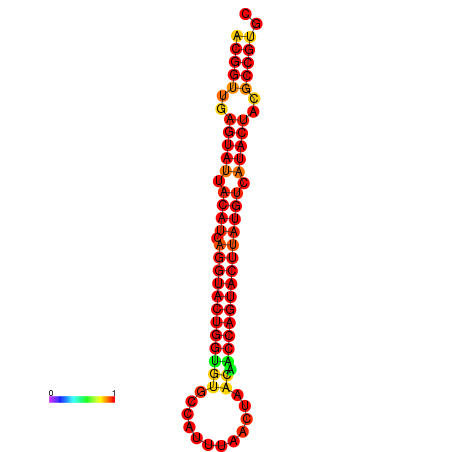 |
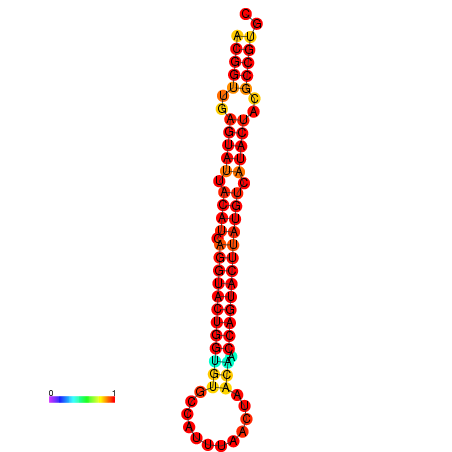 |
 |
 |
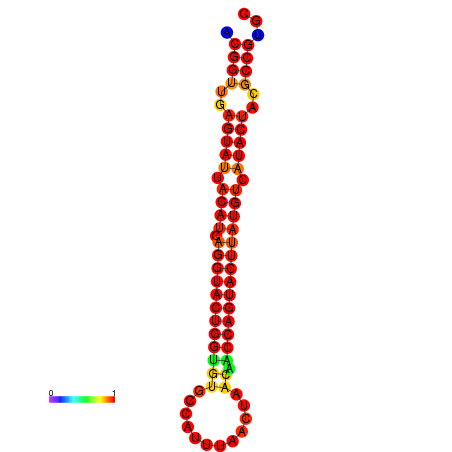 |
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
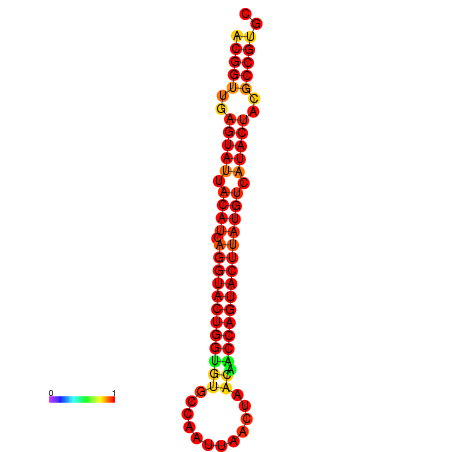 |
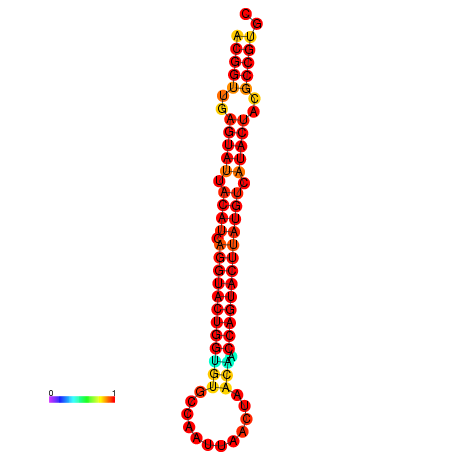 |
 |
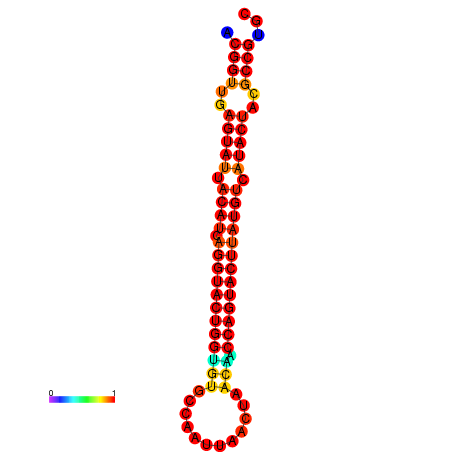 |
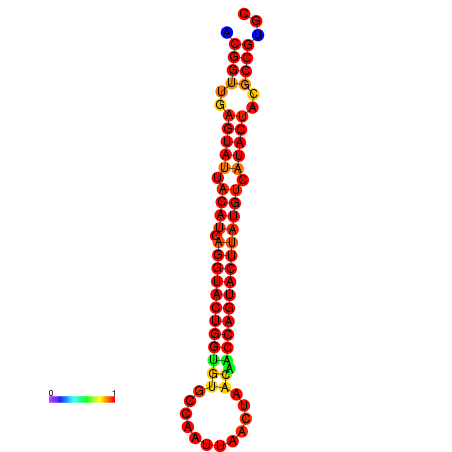 |
| dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
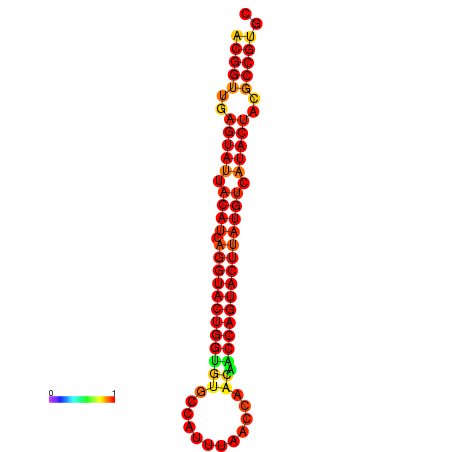 |
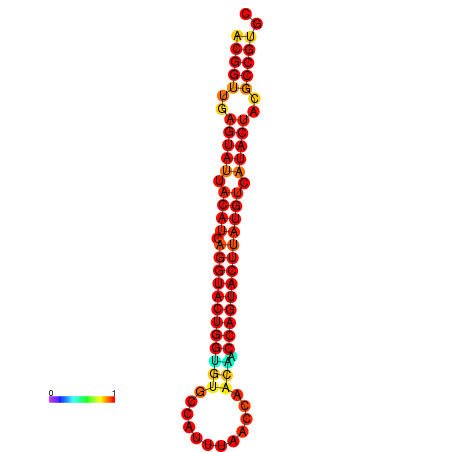 |
 |
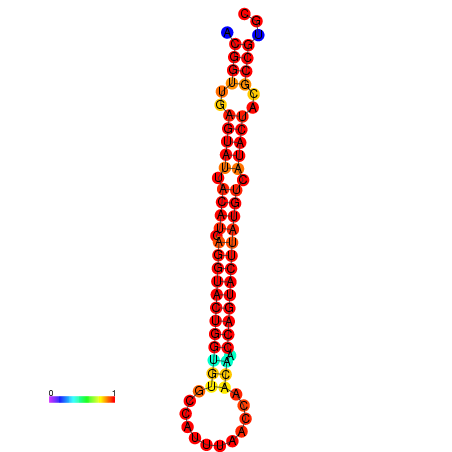 |
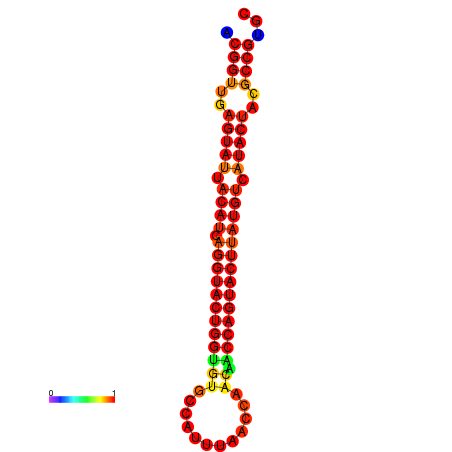 |
Generated: 03/07/2013 at 04:50 PM