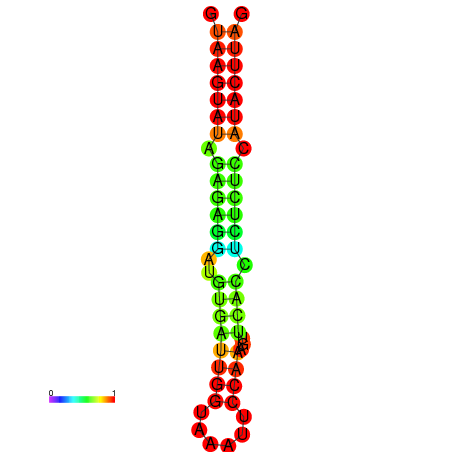
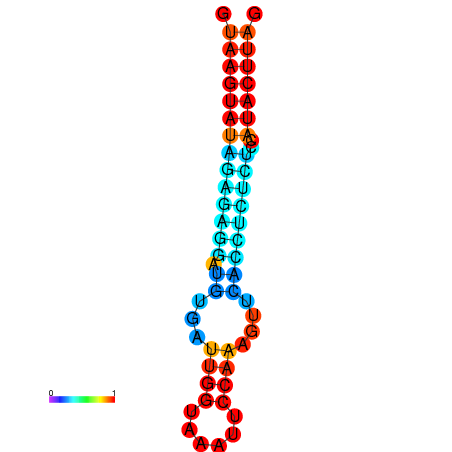
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:10120620-10120707 - | CTCCAAGCTTTTCCAGTAAGTATAGAGAGGATGTGATTGGTAAAT------TCCAAA----------GTT-----------------CACCTCTCTCCATAC-TTAGTCGTCGAGGGGTTAT |
| droSim1 | chr2R:8883298-8883385 - | CTCCAAGCTTTTCCAGTAAGTACAGAGATGATGTAATGCGTAAAT------TCCAAA----------GTT-----------------CACCTCTCTCCATAC-TTAGTCGTCGAGGGGTTAT |
| droSec1 | super_1:7628246-7628333 - | CTCCAAGCTTTTCCAGTAAGTATAGAGAGGATGTAATGCGTATAT------TCCAAT----------GTT-----------------CACCTCTCTCCATAC-TTAGTCGTCGAGGGGTTAT |
| droYak2 | chr2R:10057668-10057755 - | CTCCAAGCTGTTCCAGTAAGTATAGAGGGGATGTGATGGGTAAAT------TCCAAA----------GTT-----------------CACCTCTCTCCATAC-TTAGTCGTCGAGGGGTTAT |
| droEre2 | scaffold_4845:15723071-15723158 + | CTCCAAGCTGTTTCAGTAAGTATAGAGAGGATGTGATGGGCAAAA------TCCAAA----------GTT-----------------CACCTCTCTCCATAC-TTAGTCGTCGAGGGGTTAT |
| droAna3 | scaffold_13266:5804278-5804360 + | CTCTAAACTTTTTCAGTGAGTACTTGTAATATA-AATCCATAAA--------TCAAA----------ACT-----------------CACTT-TTTCCA-AC-TTAGTCGCCGTGGTGTGAT |
| dp4 | chr3:17369366-17369470 - | CTCAAAGCTCTTCCAGTGAGTACACAATTCAATTCCTCGGCAAGGCTATCGTCTGAAACTAATTTCCCTT-----------------CACTTATCCCTGCCCTGCAGTCGTCGCGGCGTCAT |
| droPer1 | super_31:422912-423016 + | TTCAAAGCTCTTCCAGTGAGTACACAATTCAATTCCTCGGCAAGGCTATTGTCTGAAACTAATGTCCCTT-----------------CACTTATCCCTGCCCTGCAGTCGGCGCGGCGTCAT |
| droWil1 | scaffold_180700:1839743-1839828 + | TTCCAAATTGTTCCAGTAAGTTAAAA-----TGATCTAAAATCAT------TTCAAA----------T-T----------GATT---CAATAATCTCTTCCA-TTAGTCGACGCGGTGTCAT |
| droVir3 | scaffold_12875:17586457-17586541 + | TTCCAAACTATTTCAGTAAGTAGCA------GGTGCTGAGTGCAT------G----A----TTGTCTCTA----TATCTT--------GATT-----ATCTTTGCAGTCGACGCGGGGTTAT |
| droMoj3 | scaffold_6496:11208550-11208642 + | TTCCAAGCTGTTTCAGTAAGTAGAGCACGCATTTCCAGTACCCAACTA----------------TTCGTTACTATATCTG--------TACA-----ATTTTTGTAGTCGACGCGGGGTTAT |
| droGri2 | scaffold_15245:14096515-14096602 - | TTCCAAACTGTTTCAGTAAGTGGTAACACA-ACTACTATGTTTGT------GC-AAC----------ATT----------TAATGGACAG-T-----ATCTTTGCAGTCGACGGGGAGTTAT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:10120620-10120707 - | CTCCAAGCTTTTCCAGTAAGTATAGAGAGGATGTGATTGGTAAATTCCAAAGTTCACCTCTCTCCATACTTAGTCGTCGAGGGGTTAT |
| droSim1 | chr2R:8883298-8883385 - | CTCCAAGCTTTTCCAGTAAGTACAGAGATGATGTAATGCGTAAATTCCAAAGTTCACCTCTCTCCATACTTAGTCGTCGAGGGGTTAT |
| droSec1 | super_1:7628246-7628333 - | CTCCAAGCTTTTCCAGTAAGTATAGAGAGGATGTAATGCGTATATTCCAATGTTCACCTCTCTCCATACTTAGTCGTCGAGGGGTTAT |
| droYak2 | chr2R:10057668-10057755 - | CTCCAAGCTGTTCCAGTAAGTATAGAGGGGATGTGATGGGTAAATTCCAAAGTTCACCTCTCTCCATACTTAGTCGTCGAGGGGTTAT |
| droEre2 | scaffold_4845:15723071-15723158 + | CTCCAAGCTGTTTCAGTAAGTATAGAGAGGATGTGATGGGCAAAATCCAAAGTTCACCTCTCTCCATACTTAGTCGTCGAGGGGTTAT |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
| dG=-20.9, p-value=0.009901 | dG=-20.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-9.8, p-value=0.009901 | dG=-9.2, p-value=0.009901 | dG=-8.9, p-value=0.009901 |
|---|---|---|
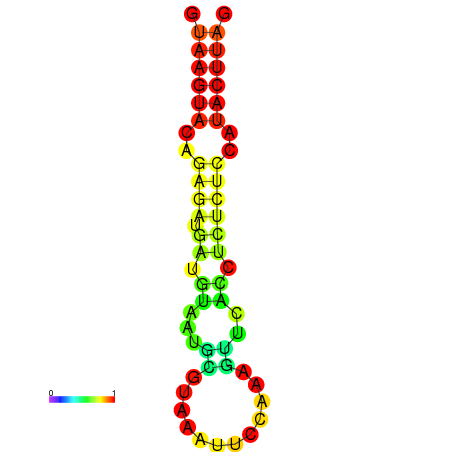 |
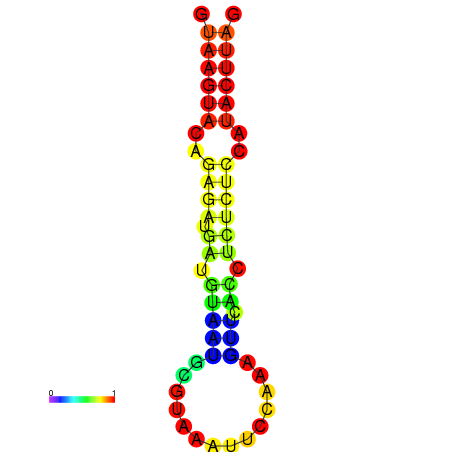 |
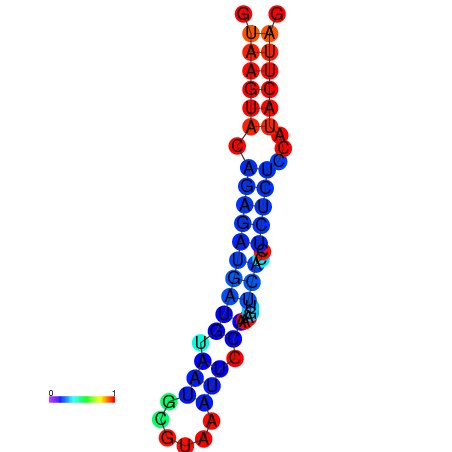 |
| dG=-16.5, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.5, p-value=0.009901 |
|---|---|---|
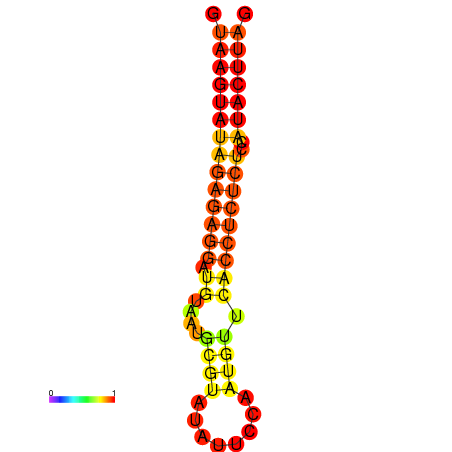 |
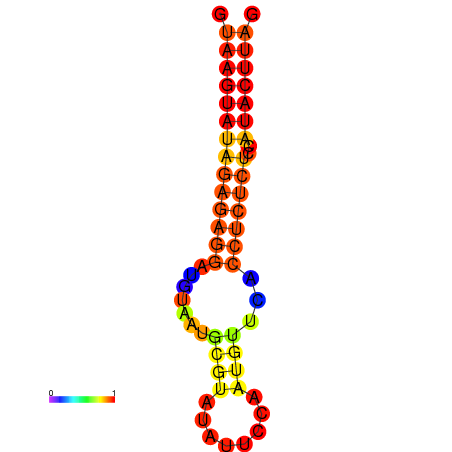 |
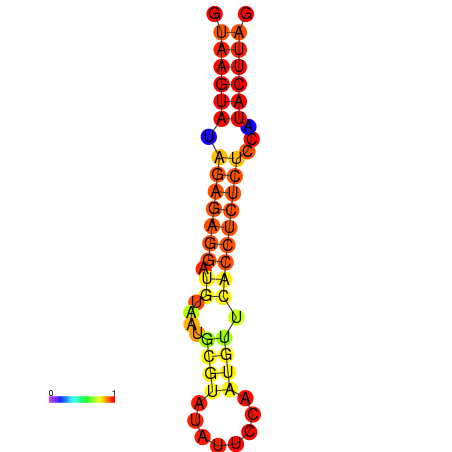 |
| dG=-19.3, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|
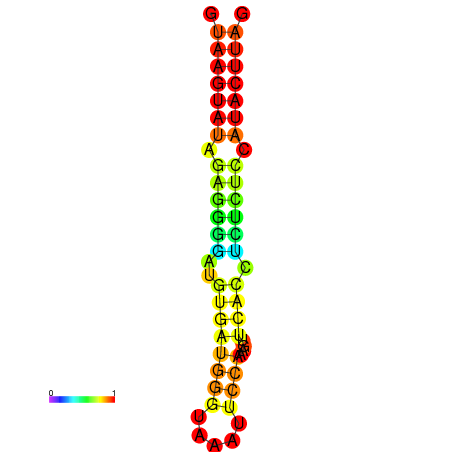 |
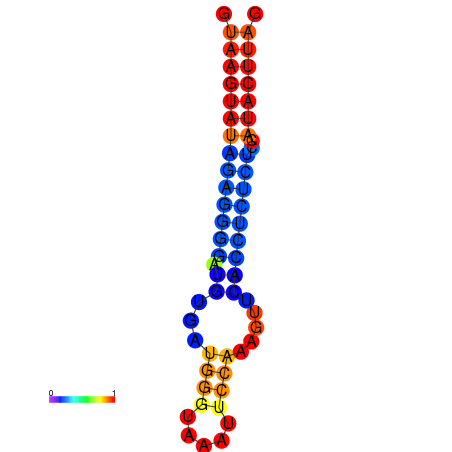 |
| dG=-19.3, p-value=0.009901 | dG=-18.9, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|---|
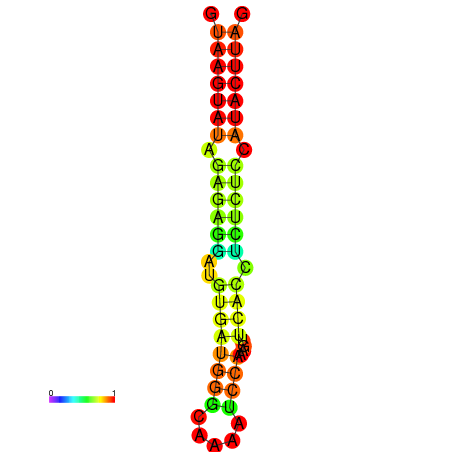 |
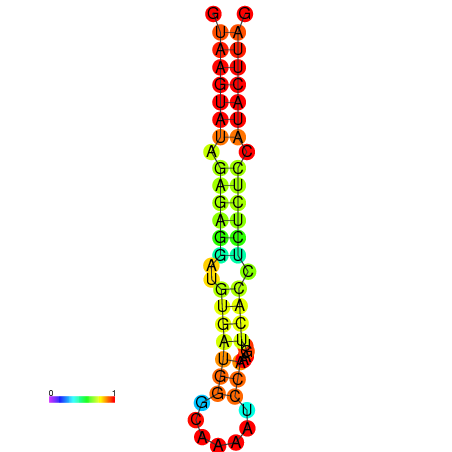 |
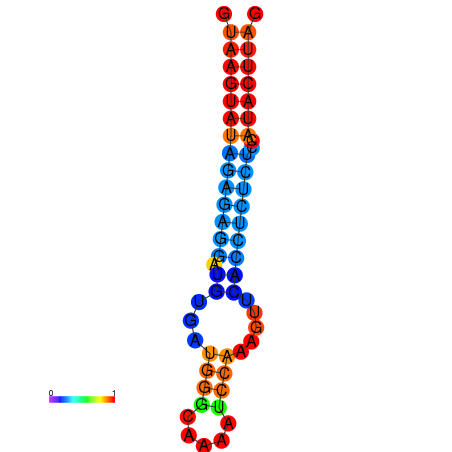 |
| dG=-6.1, p-value=0.009901 | dG=-5.4, p-value=0.009901 |
|---|---|
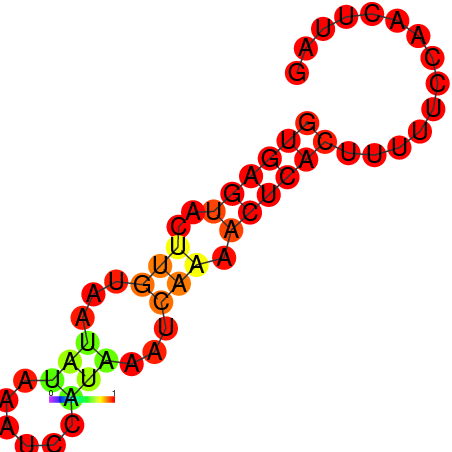 |
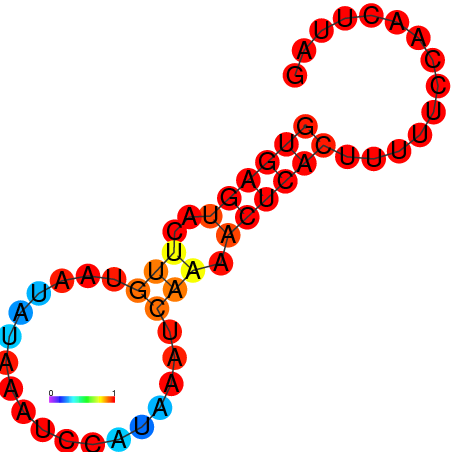 |
| dG=-9.5, p-value=0.009901 | dG=-9.1, p-value=0.009901 | dG=-8.74, p-value=0.009901 | dG=-8.5, p-value=0.009901 | dG=-7.6, p-value=0.009901 |
|---|---|---|---|---|
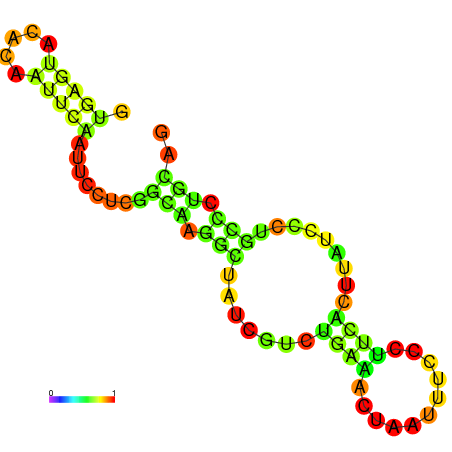 |
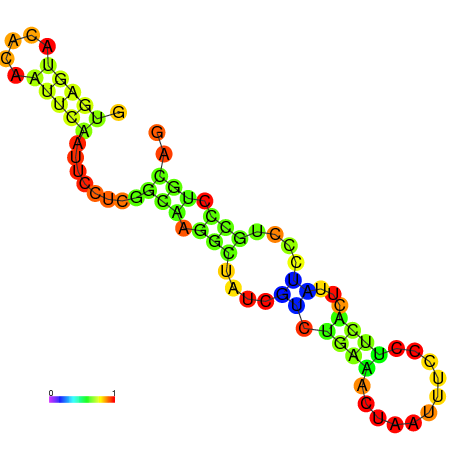 |
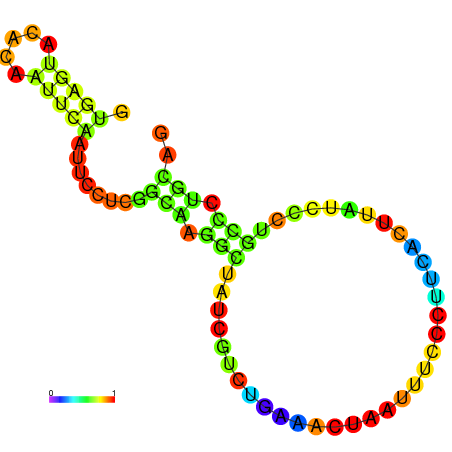 |
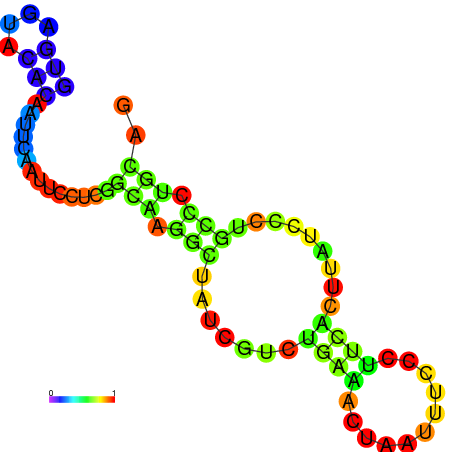 |
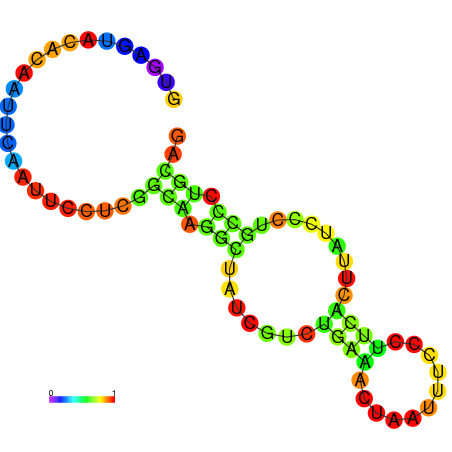 |
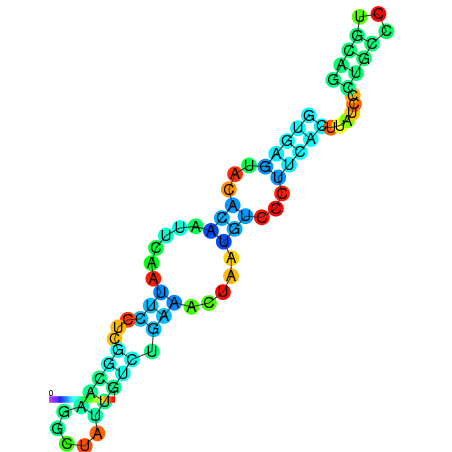
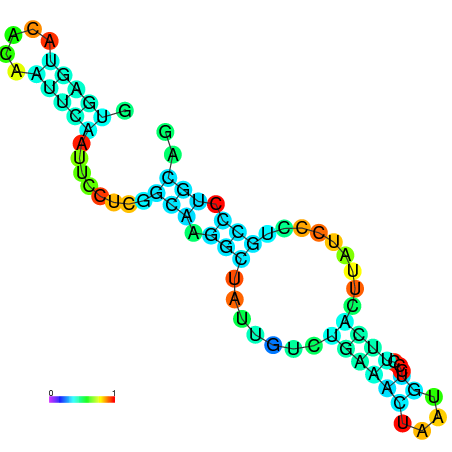
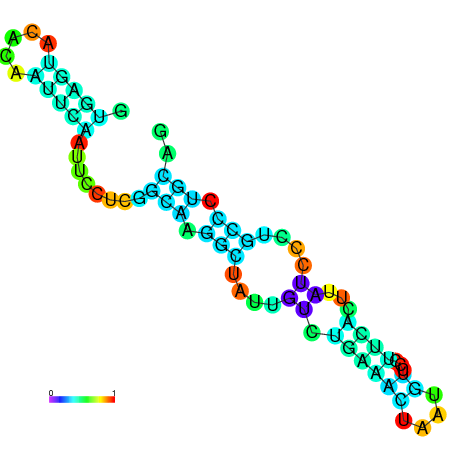
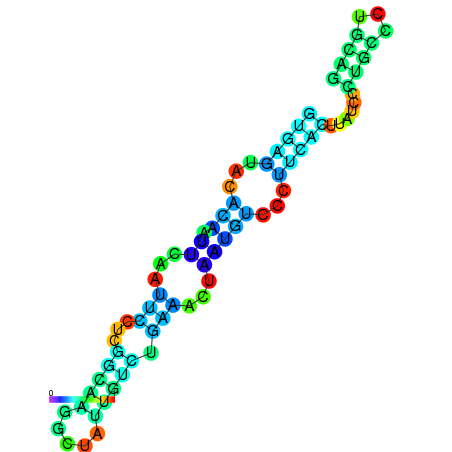
| dG=-10.2, p-value=0.009901 | dG=-10.1, p-value=0.009901 | dG=-9.7, p-value=0.009901 | dG=-9.6, p-value=0.009901 | dG=-9.6, p-value=0.009901 | dG=-8.2, p-value=0.009901 |
|---|---|---|---|---|---|
 |
 |
 |
 |
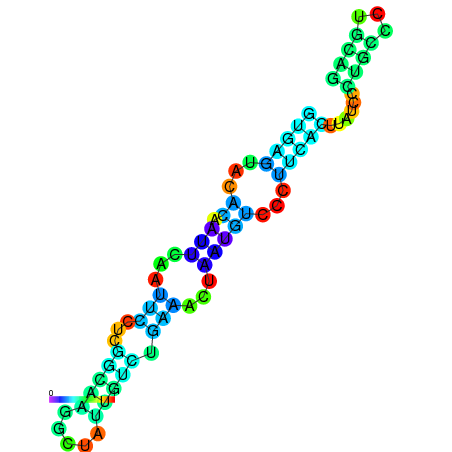 |
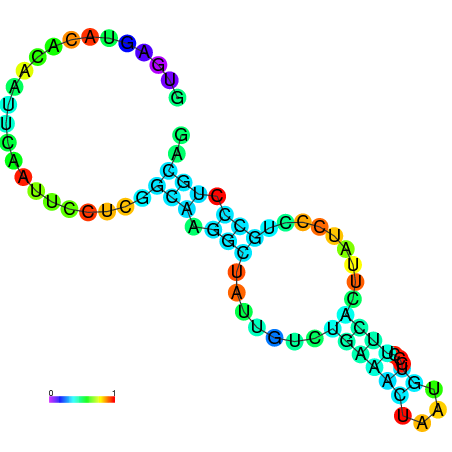 |
| dG=-3.5, p-value=0.009901 | dG=-3.0, p-value=0.009901 | dG=-2.8, p-value=0.009901 | dG=-2.8, p-value=0.009901 | dG=-2.7, p-value=0.009901 |
|---|---|---|---|---|
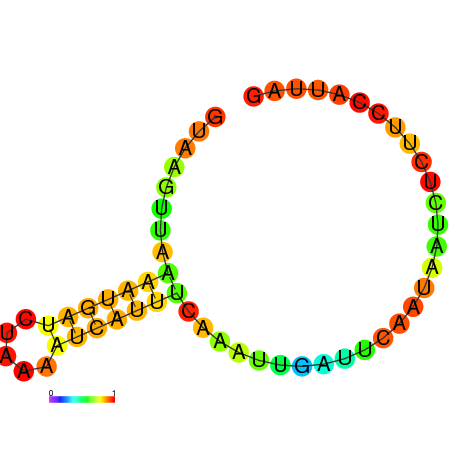 |
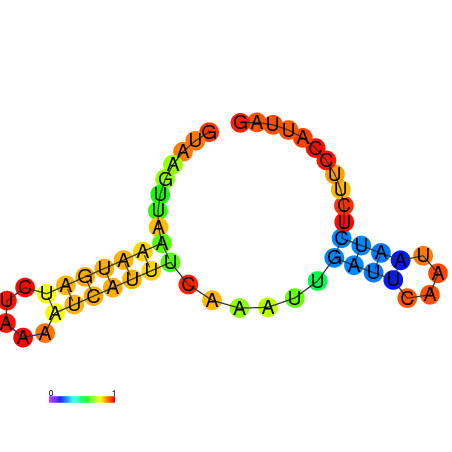 |
 |
 |
 |
| dG=-9.5, p-value=0.009901 | dG=-9.1, p-value=0.009901 | dG=-8.9, p-value=0.009901 | dG=-8.8, p-value=0.009901 | dG=-8.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-1.7, p-value=0.009901 | dG=-1.6, p-value=0.019802 | dG=-1.6, p-value=0.009901 | dG=-1.6, p-value=0.009901 | dG=-1.5, p-value=0.009901 | dG=-1.4, p-value=0.009901 |
|---|---|---|---|---|---|
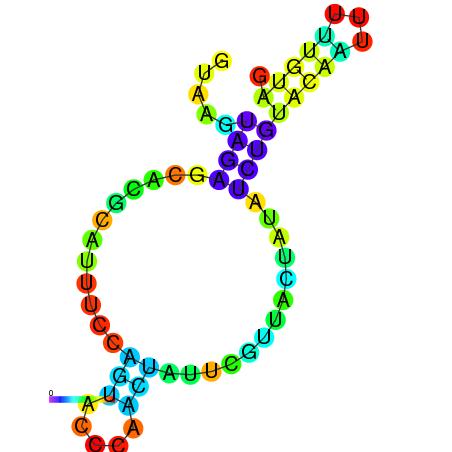 |
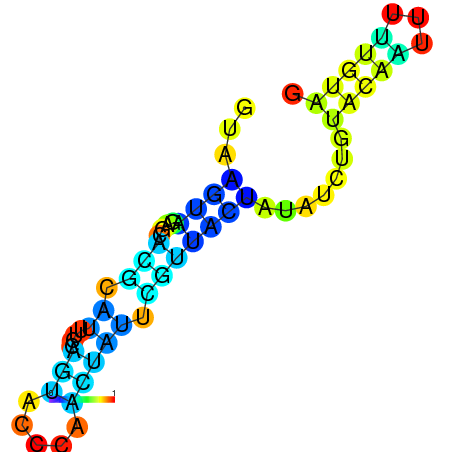 |
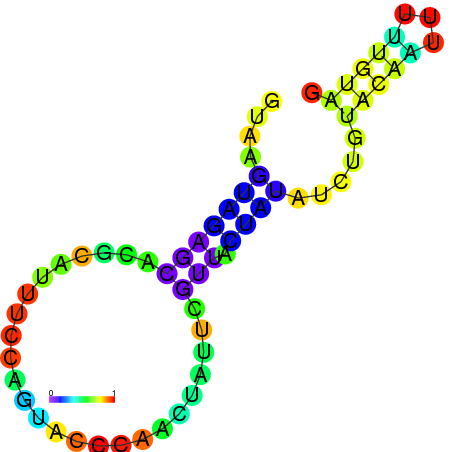 |
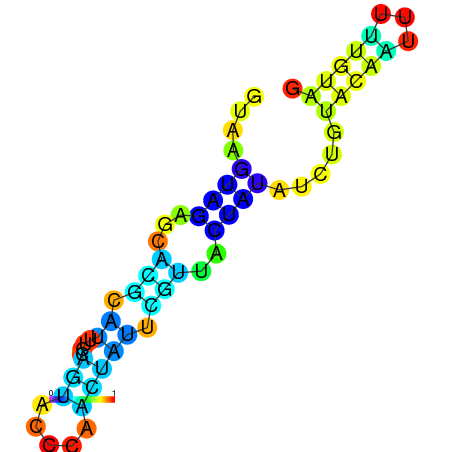 |
 |
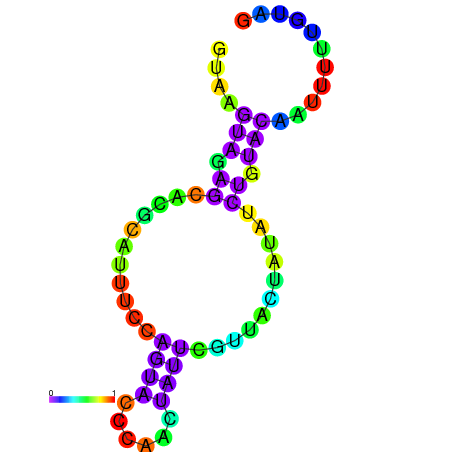 |
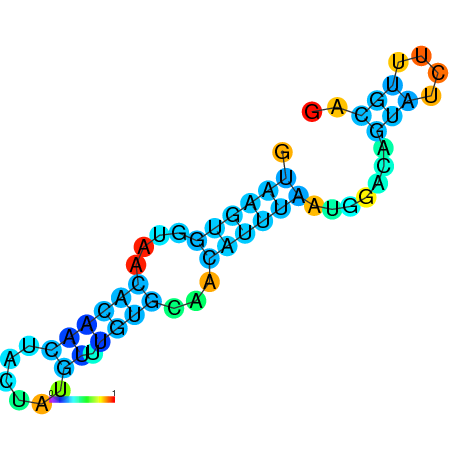
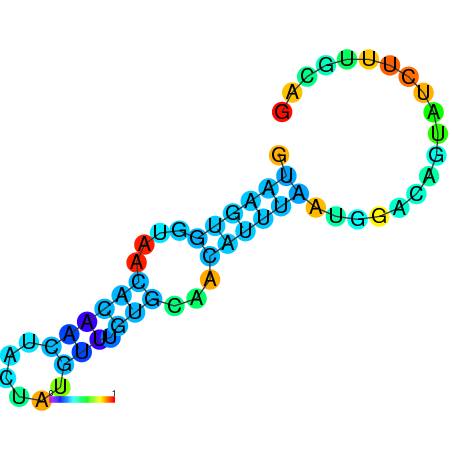
| dG=-8.2, p-value=0.009901 | dG=-8.0, p-value=0.009901 | dG=-8.0, p-value=0.009901 | dG=-7.8, p-value=0.009901 | dG=-7.8, p-value=0.009901 | dG=-7.6, p-value=0.009901 |
|---|---|---|---|---|---|
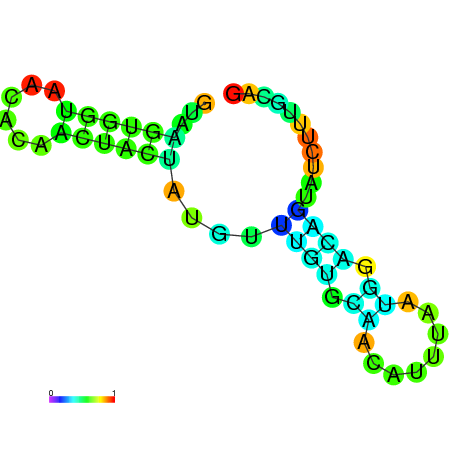 |
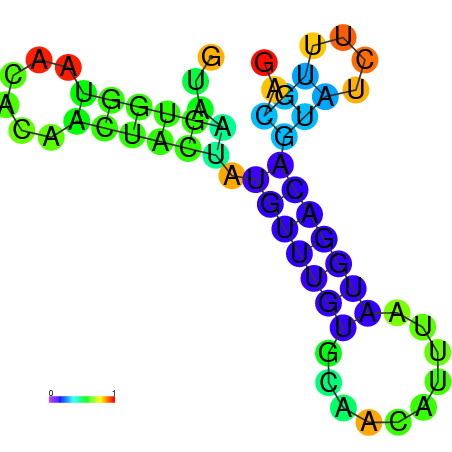 |
 |
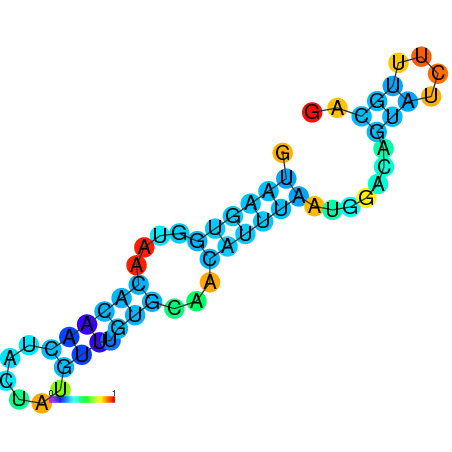 |
 |
 |
Generated: 03/07/2013 at 04:48 PM