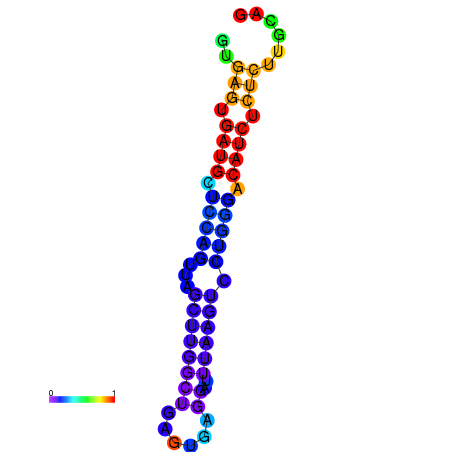
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:20164939-20165032 + | TCATGGGCTACGACGGTGAGTG-----ATGCTCCAGTTAGCTTGGCTGAGTG-------AGGATTTA------------AGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAAG |
| droSim1 | chr3R:19998542-19998635 + | TCATGGGCTACGACGGTGAGTG-----ATGCTCCAGTTAGCTTGGCTGAGTG-------AGGATTTA------------AGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droSec1 | super_0:20503496-20503589 + | TCATGGGCTACGACGGTGAGTG-----ATGCTCCAGTTAGCTTGTCTGAGTG-------AGACTTTA------------AGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droYak2 | chr3R:26340571-26340664 - | TCATGGGCTACGACGGTGAGTG-----ATGCTCCAGATAGCTTGACTGGGTG-------AGGATTTA------------AGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAAC |
| droEre2 | scaffold_4820:7895693-7895786 - | TCATGGGCTACGACGGTGAGTG-----ATGCTCCAGATAGCTTGGCTGAGTG-------AGGACTTA------------AGTCCTGGGAC-----ATCTCTCTTGCAGTCAAAATCCTAAAAC |
| droAna3 | scaffold_13088:204245-204339 + | TCATGGGCTACGACAGTGAGTG-----ATGCTCCAGATAGGTTGGCTTAGGAAGTC------ATTAA------------AGCCCTGGGAT-----GTCTCTCTTGCAGTTAAGATCCTGAAAC |
| dp4 | chr2:18646206-18646298 - | TGATGGGCTACGATGGTAAGAG--C---TAGC---------C-----ACACAAATTATTAGTATT----------TCTATTTCTCAAAATATTT-CTTTCTTTTGCAGTCAAGATACTAAAGC |
| droPer1 | super_3:1376406-1376498 - | TGATGGGCTACGATGGTAAGAG--C---TAGC---------C-----ACACAAATTATTAGTATT----------TCTATTTCTCAAAATATTC-CTTTCTTTTGCAGTCAAGATCCTAAAGC |
| droWil1 | scaffold_181108:4541725-4541808 - | TAATGGGTTATGACAGTAAGTTAGC---TGGTAAAGTTGATTTCT------------------TTTA------------ACTAATGT-GT-----ATATTTACTGTAGTAAGAATTTTAAAGC |
| droVir3 | scaffold_13047:3497490-3497590 + | TTGTGGGCTACGAAAGTAAGTA------TTTACTTGATAACTCG------TT-------AAACCTAAAG--TCTGTTAACCACTCTTATAACTATG-CATCTAAACAGTTAAGGTACTAAAGA |
| droMoj3 | scaffold_6540:20508341-20508431 + | TAATGGGCTACGATGGTAAATG--CTGATGTTCATGAATATTTATCCAAGTA-------ATTT------------------CTTTACGTT-----GTCTTCTTAACAGTTAAGGTTTTGAAAA |
| droGri2 | scaffold_14906:156637-156731 + | TGGTGGGCTATGATAGTGAGTT--G--ATTCTATT---------------GTAATAATCA------AATCGTTTTTGTTATTTTATAAACA---TGCCATTTTAACAGTCAAGATACTCAAGA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:20164939-20165032 + | TCATGGGCTACGACGGTGAGTGATGCTCCAGTTAGCTTGGCTGAGTGAGG-ATTTAAGTCCTGGGACATCTCTCTTGCAGTCAAGATCCTGAAAG |
| droSim1 | chr3R:19998542-19998635 + | TCATGGGCTACGACGGTGAGTGATGCTCCAGTTAGCTTGGCTGAGTGAGG-ATTTAAGTCCTGGGACATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droSec1 | super_0:20503496-20503589 + | TCATGGGCTACGACGGTGAGTGATGCTCCAGTTAGCTTGTCTGAGTGAGA-CTTTAAGTCCTGGGACATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droYak2 | chr3R:26340571-26340664 - | TCATGGGCTACGACGGTGAGTGATGCTCCAGATAGCTTGACTGGGTGAGG-ATTTAAGTCCTGGGACATCTCTCTTGCAGTCAAGATCCTGAAAC |
| droEre2 | scaffold_4820:7895693-7895786 - | TCATGGGCTACGACGGTGAGTGATGCTCCAGATAGCTTGGCTGAGTGAGG-ACTTAAGTCCTGGGACATCTCTCTTGCAGTCAAAATCCTAAAAC |
| droAna3 | scaffold_13088:204245-204339 + | TCATGGGCTACGACAGTGAGTGATGCTCCAGATAGGTTGGCTTAGGAAGTCATTAAAGCCCTGGGATGTCTCTCTTGCAGTTAAGATCCTGAAAC |
| Species | Read pileup | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-18.8, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-17.9, p-value=0.009901 | dG=-17.9, p-value=0.009901 | dG=-17.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-18.8, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-17.9, p-value=0.009901 | dG=-17.9, p-value=0.009901 | dG=-17.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
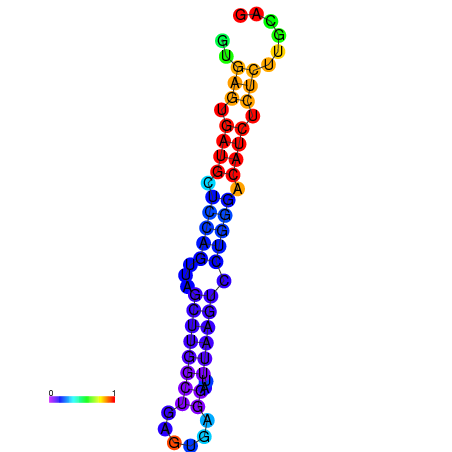 |
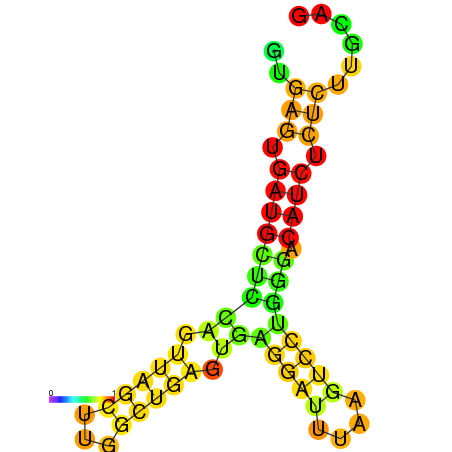
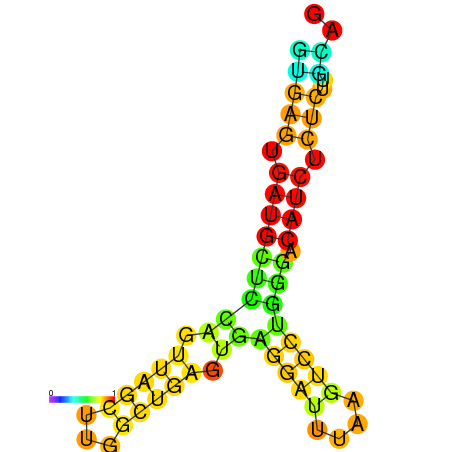
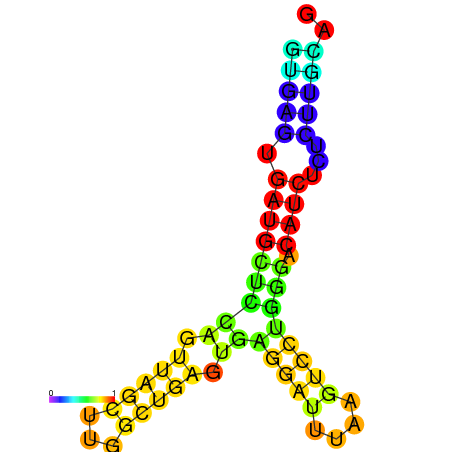
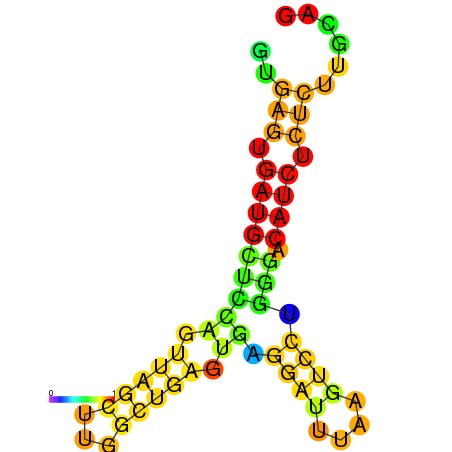
| dG=-20.0, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.1, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-17.8, p-value=0.009901 | dG=-17.7, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.3, p-value=0.009901 | dG=-17.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-18.9, p-value=0.009901 | dG=-18.9, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
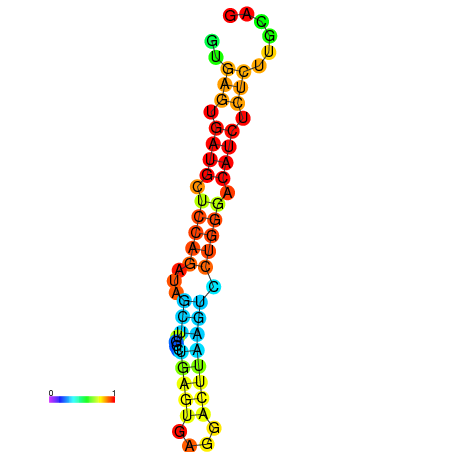 |
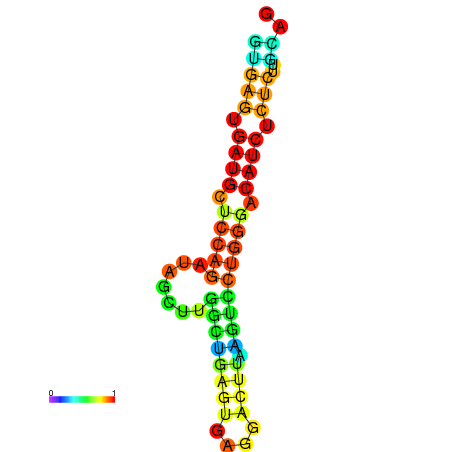
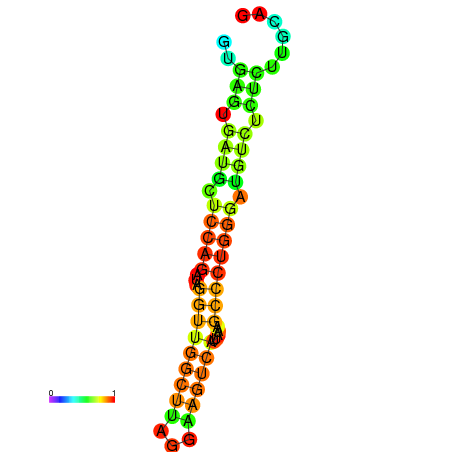
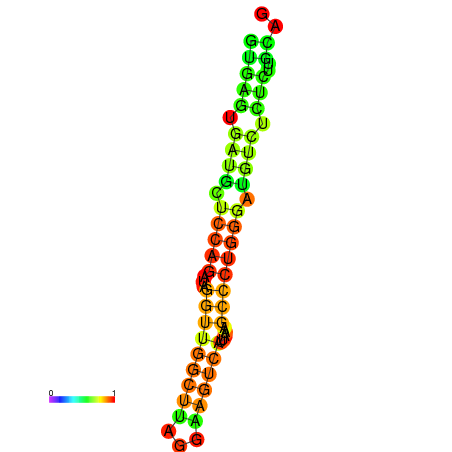
| dG=-16.7, p-value=0.009901 | dG=-16.3, p-value=0.009901 | dG=-16.3, p-value=0.009901 | dG=-16.2, p-value=0.009901 | dG=-15.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
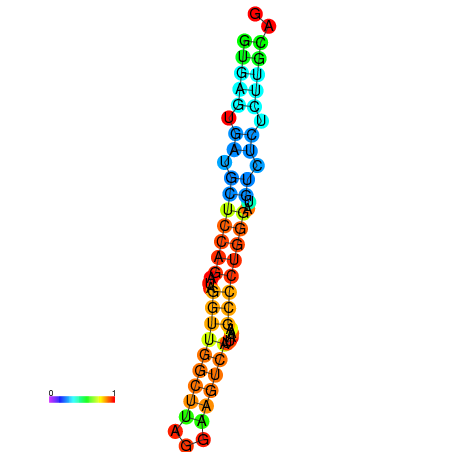 |
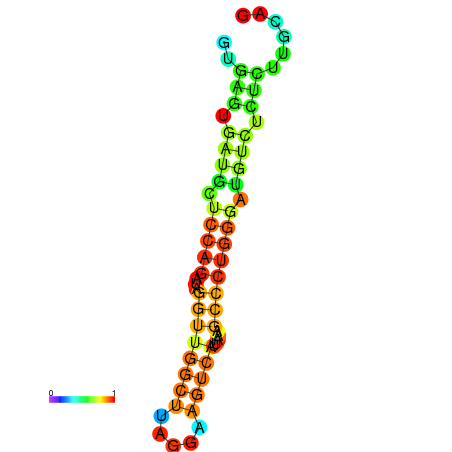 |
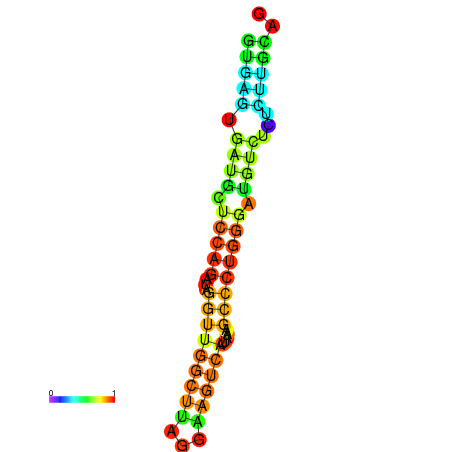 |
| dG=-4.9, p-value=0.009901 | dG=-4.2, p-value=0.009901 |
|---|---|
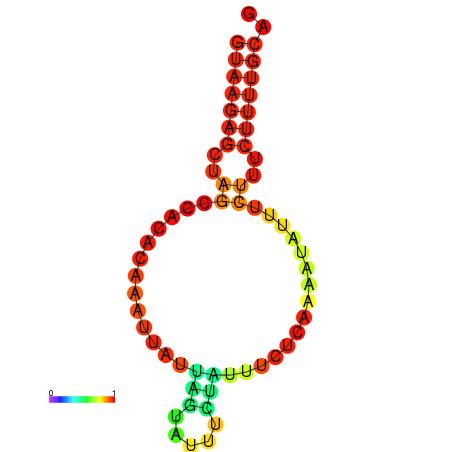 |
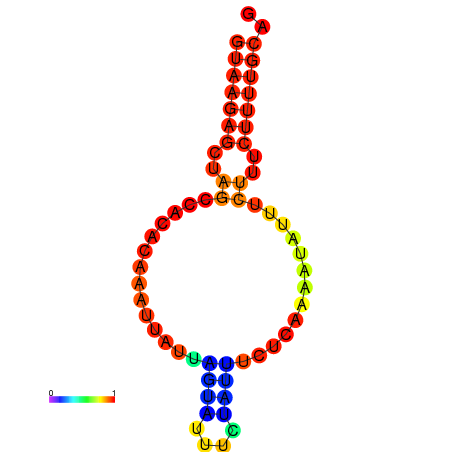 |
| dG=-4.9, p-value=0.009901 | dG=-4.2, p-value=0.009901 |
|---|---|
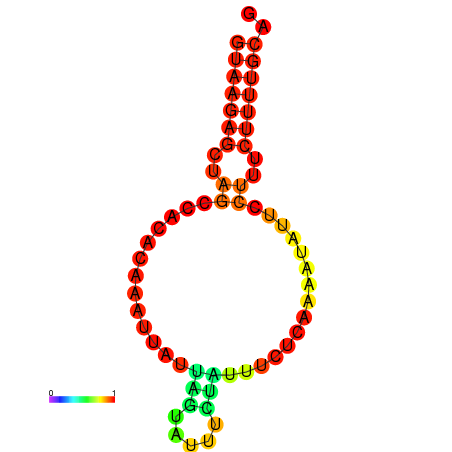 |
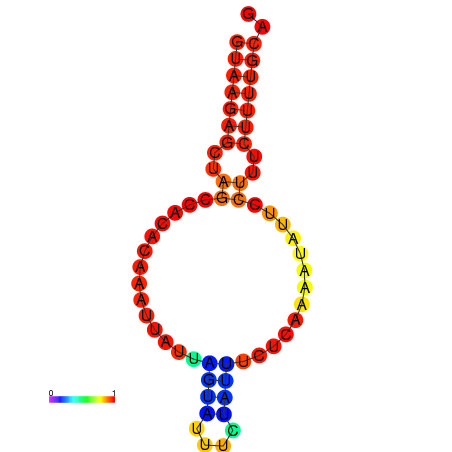 |
| dG=-8.3, p-value=0.009901 | dG=-7.6, p-value=0.009901 | dG=-7.3, p-value=0.009901 |
|---|---|---|
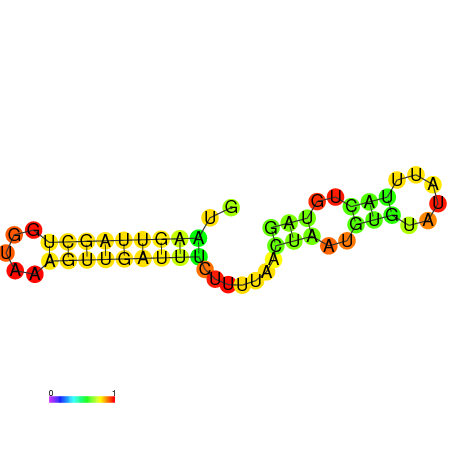 |
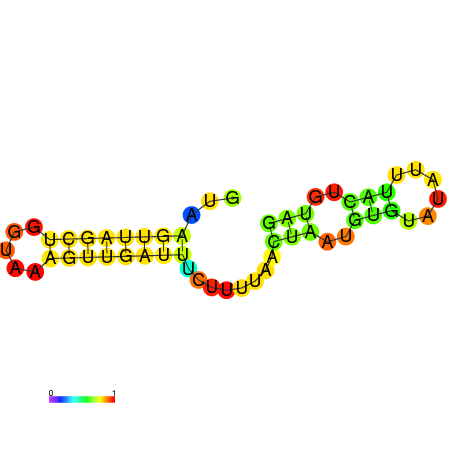 |
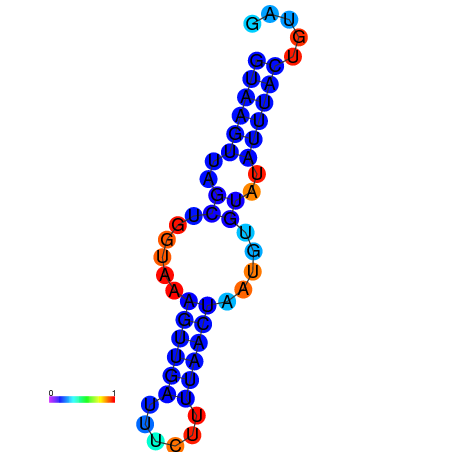 |
| dG=-2.5, p-value=0.009901 | dG=-2.3, p-value=0.009901 | dG=-1.5, p-value=0.009901 |
|---|---|---|
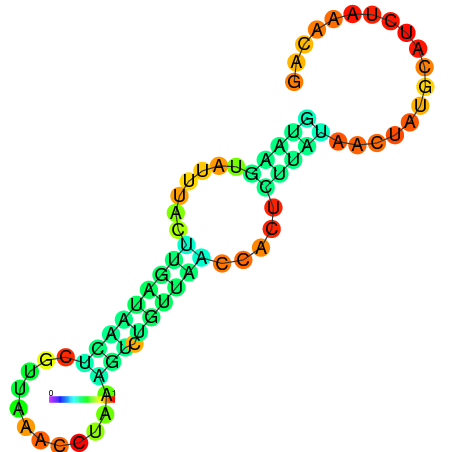 |
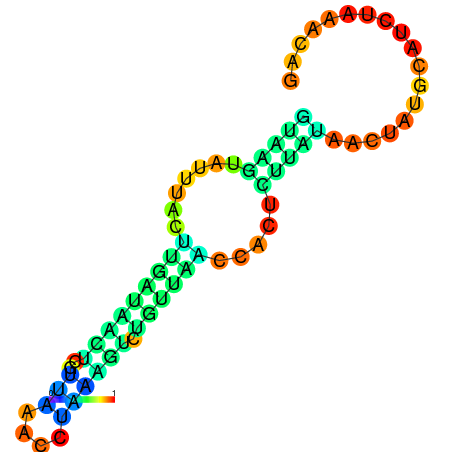 |
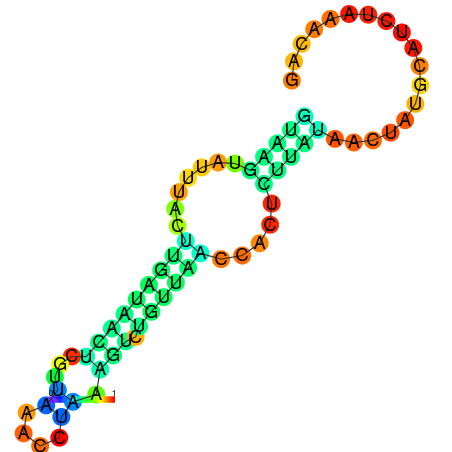 |
| dG=-3.5, p-value=0.009901 | dG=-2.9, p-value=0.009901 | dG=-2.9, p-value=0.009901 | dG=-2.7, p-value=0.009901 | dG=-2.6, p-value=0.009901 |
|---|---|---|---|---|
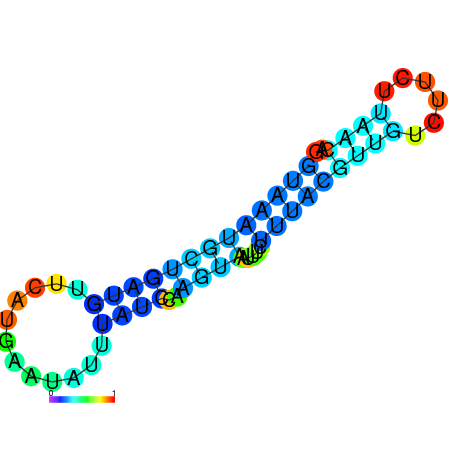 |
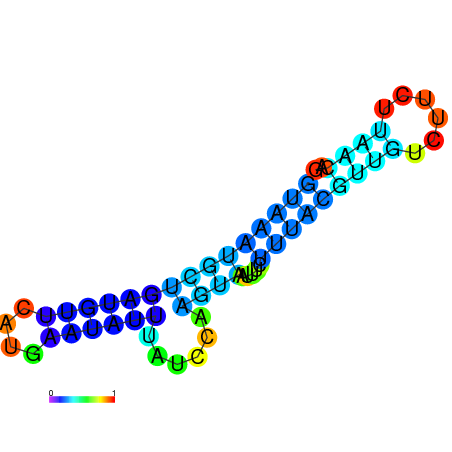 |
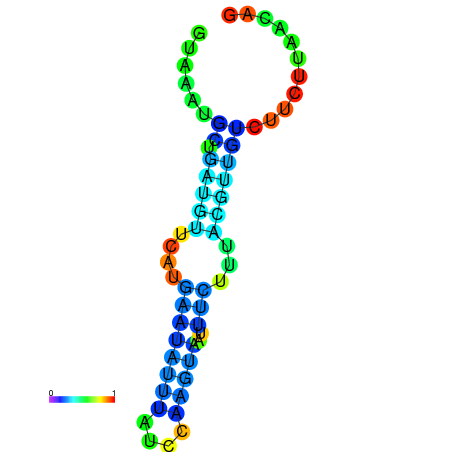 |
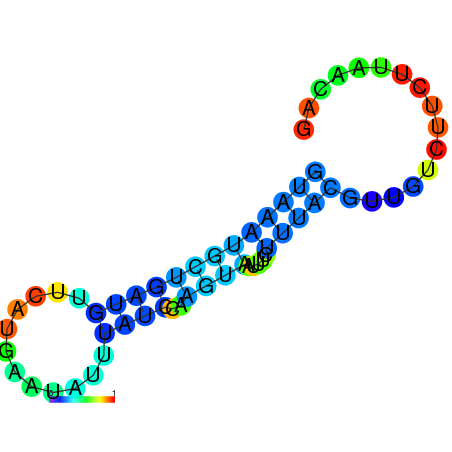 |
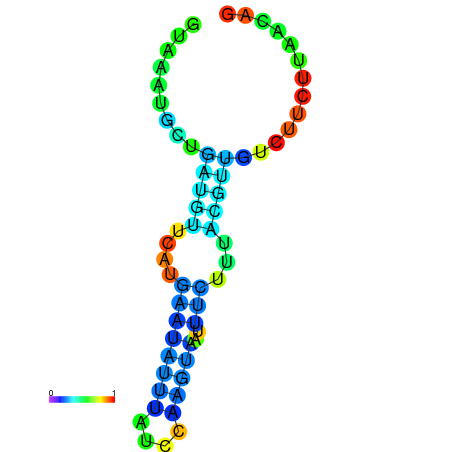 |
| dG=-3.9, p-value=0.009901 | dG=-3.8, p-value=0.009901 | dG=-3.8, p-value=0.009901 | dG=-3.5, p-value=0.009901 | dG=-3.5, p-value=0.009901 |
|---|---|---|---|---|
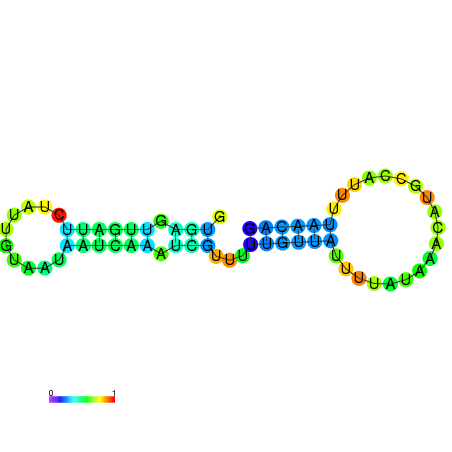 |
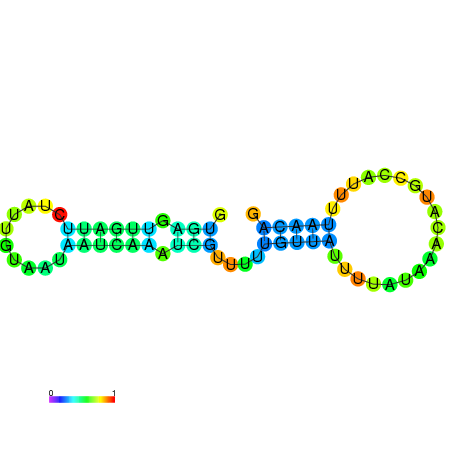 |
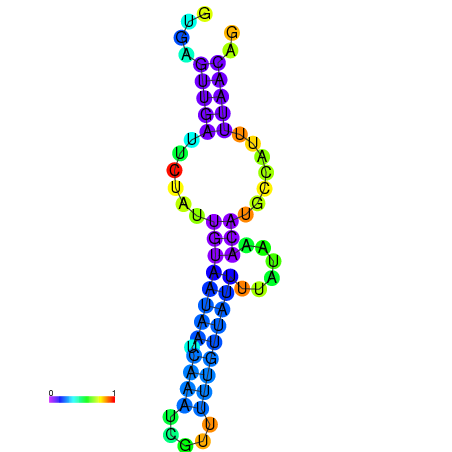 |
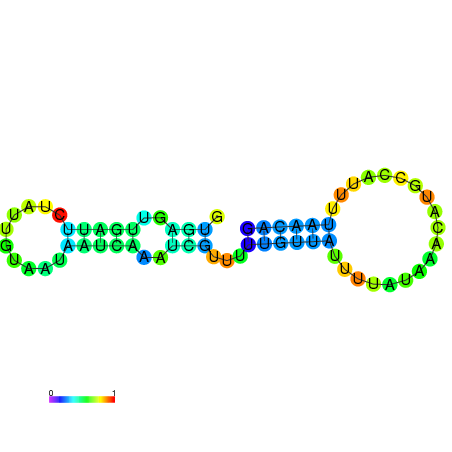 |
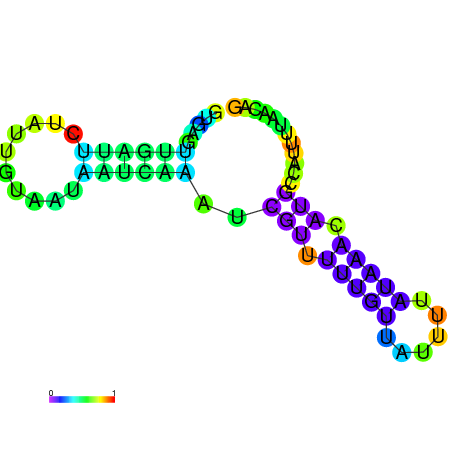 |
Generated: 03/07/2013 at 04:48 PM