
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:27579231-27579328 - | TGGCCTTTTCTCAAGGTATAATGGAAATAGATTTTAAT------CGCAGGC--------------GCGTCAGTGGT---------------TGA--ATTAAAATTCATTTTCATTTGCAGTTACTGGATACAACA |
| droSim1 | chr3R:27196220-27196317 - | TGGCCTTCTCCCAAGGTATAATGGAAATAGATTTTAAT------CGCTGGC--------------GCGTCAGTGGT---------------TGA--ATTAAAATTCATTTTCATTTGCAGTTACCGGATACAACA |
| droSec1 | super_31:233822-233919 - | TGGCCTTCTCCCAAGGTATAATGGAAATAGATTTTAAT------CGCTGGC--------------GCGTCAGTGGT---------------TGA--ATTAAAATTCATTTTCATTTGCAGTTACCGGATACAACA |
| droYak2 | chr3R:28513868-28513965 - | TGGCCTTTTCCCAAGGTATAATGGAAATAGATTTTAAT------CGCAGGC--------------GCGTCAGTGGT---------------TGA--ATTAAAATTCATTTTCATTTGCAGTCACCGGATACAACA |
| droEre2 | scaffold_4820:396401-396498 + | TGGCCTTTTCGCAAGGTATAATGGAAATAGATTTTAAT------CGCAGGC--------------GCGTCAGTGGT---------------TGA--ATTAAAATTCATTTTCATTTGCAGTCACCGGATACAACA |
| droAna3 | scaffold_12911:3510618-3510715 + | TGGCATTCTCCCAAGGTATAATGAAAATTGATTTTAAT------CACACGG--------------ATCGGAGTGGC---------------AAA--ATTAAAATTCATTTTCATTTGCAGTCACGGGATACAATG |
| dp4 | chr2:14711225-14711322 + | TGGCCTTCTCCCAAGGTACAATGGAAATAGATTTTAAT------CGGGTTT--------------CGTTTGGCGGT---------------GAA--ATTAAAATTCATTTTCATTTACAGTGACCGGCATCAACA |
| droPer1 | super_0:5703122-5703219 - | TGGCCTTCTCCCAAGGTACAATGGAAATAGATTTTAAT------CGTGTTT--------------CGTTTGGCGGT---------------GAA--ATTAAAATTCATTTTCATTTACAGTGACCGGCATCAACA |
| droWil1 | scaffold_181130:8196915-8197021 + | TGGCATTTTCTCAAGGTGAAATGAAAATAAATTTTAATTCGGATAGAAAGCTAAT--CTAAT--------TGT-AT---------------TGC--ATTAAAATTCGTTTTCATTTGCAGTATCTGGTCTAAATA |
| droVir3 | Unknown | TGGCATTCTCTCAAGGTAT---GTACATATTTTTAAAT-----------------------------------------------------TTA--ACCAGTATTTACTTTTGTTCACAGTGACCGGCATAAATA |
| droMoj3 | scaffold_6540:30646887-30646982 + | TTGCCTTCTCTCAAGGTAAG---TGC------TTTGACTTAGCA--AAAGTTATTTACTAGAAGTGTTTCTCTGC----------------------------TTTCGTTTCATCCACAGTGACCGGTGTAAATA |
| droGri2 | scaffold_14624:997050-997159 - | TGGCCTTTTCACAGGGTATGCAGAGCTTG-ATTAT--T------TGATTGC--------------TCATTT--TACACATTATAAATAAATTATATATATATATACATATGTTATTGCAGTGACTGGCGTAAATA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:27579231-27579328 - | TGGCCTTTTCTCAAGGTATAATGGAAATAGATTTTAATC------GCAGGCGCGTC----AGTGGTTGAATTAAAATTCATTTTCATTTGCAGTTACTGGATACAACA |
| droSim1 | chr3R:27196220-27196317 - | TGGCCTTCTCCCAAGGTATAATGGAAATAGATTTTAATC------GCTGGCGCGTC----AGTGGTTGAATTAAAATTCATTTTCATTTGCAGTTACCGGATACAACA |
| droSec1 | super_31:233822-233919 - | TGGCCTTCTCCCAAGGTATAATGGAAATAGATTTTAATC------GCTGGCGCGTC----AGTGGTTGAATTAAAATTCATTTTCATTTGCAGTTACCGGATACAACA |
| droYak2 | chr3R:28513868-28513965 - | TGGCCTTTTCCCAAGGTATAATGGAAATAGATTTTAATC------GCAGGCGCGTC----AGTGGTTGAATTAAAATTCATTTTCATTTGCAGTCACCGGATACAACA |
| droEre2 | scaffold_4820:396401-396498 + | TGGCCTTTTCGCAAGGTATAATGGAAATAGATTTTAATC------GCAGGCGCGTC----AGTGGTTGAATTAAAATTCATTTTCATTTGCAGTCACCGGATACAACA |
| droAna3 | scaffold_12911:3510618-3510715 + | TGGCATTCTCCCAAGGTATAATGAAAATTGATTTTAATC------ACACGGATCGG----AGTGGCAAAATTAAAATTCATTTTCATTTGCAGTCACGGGATACAATG |
| dp4 | chr2:14711225-14711322 + | TGGCCTTCTCCCAAGGTACAATGGAAATAGATTTTAATC------GGGTTTCGTTT----GGCGGTGAAATTAAAATTCATTTTCATTTACAGTGACCGGCATCAACA |
| droPer1 | super_0:5703122-5703219 - | TGGCCTTCTCCCAAGGTACAATGGAAATAGATTTTAATC------GTGTTTCGTTT----GGCGGTGAAATTAAAATTCATTTTCATTTACAGTGACCGGCATCAACA |
| droWil1 | scaffold_181130:8196915-8197021 + | TGGCATTTTCTCAAGGTGAAATGAAAATAAATTTTAATTCGGATAGAAAGCTAATCTAATTGT-ATTGCATTAAAATTCGTTTTCATTTGCAGTATCTGGTCTAAATA |
| Species | Read pileup | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||
| dp4 |
|
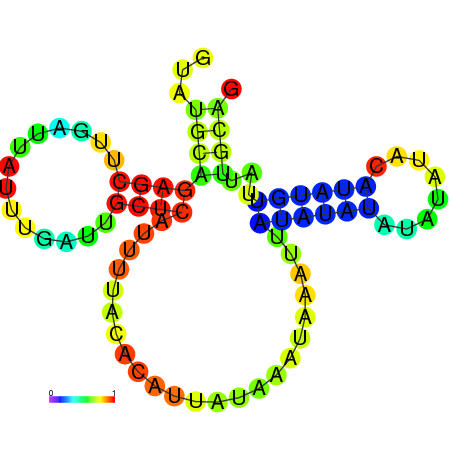
| dG=-19.7, p-value=0.009901 | dG=-18.7, p-value=0.009901 |
|---|---|
 |
 |
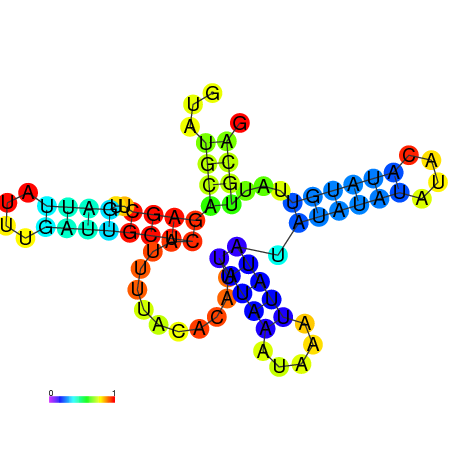
| dG=-18.2, p-value=0.009901 | dG=-18.2, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
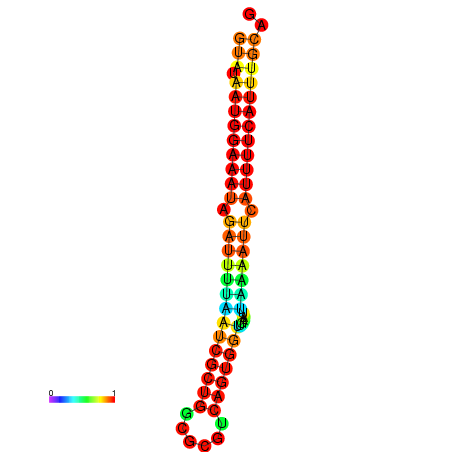 |
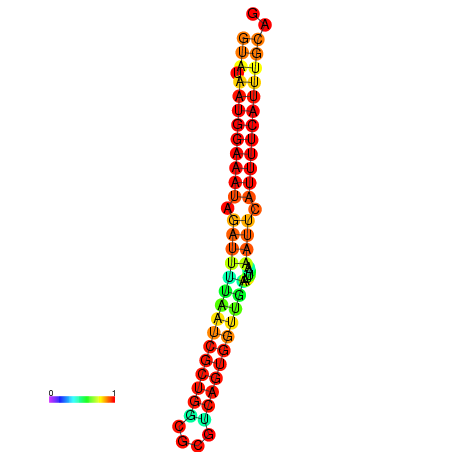
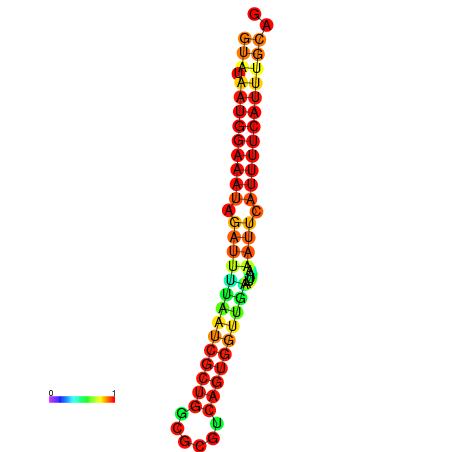
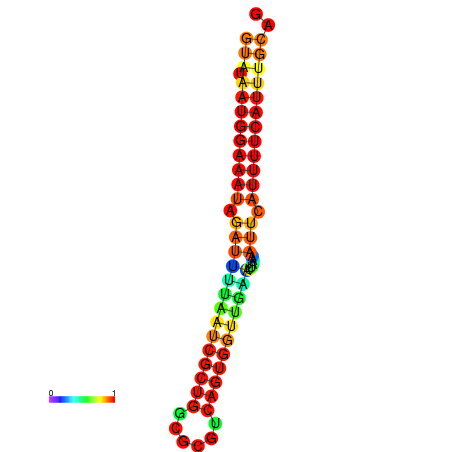
| dG=-18.2, p-value=0.009901 | dG=-18.2, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-19.7, p-value=0.009901 | dG=-18.7, p-value=0.009901 |
|---|---|
 |
 |
| dG=-19.7, p-value=0.009901 | dG=-18.7, p-value=0.009901 |
|---|---|
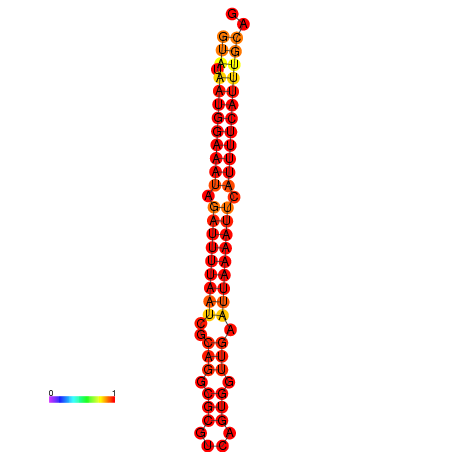 |
 |
| dG=-16.0, p-value=0.009901 | dG=-15.0, p-value=0.009901 |
|---|---|
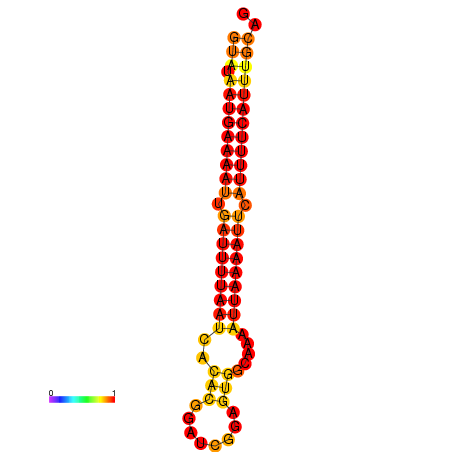 |
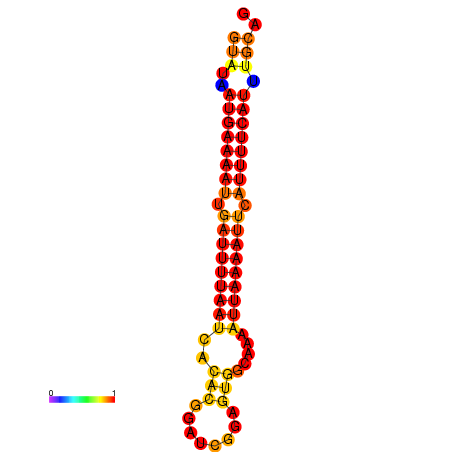 |
| dG=-14.0, p-value=0.009901 | dG=-13.5, p-value=0.009901 |
|---|---|
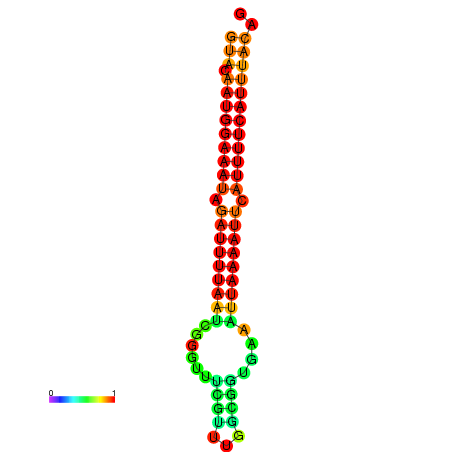 |
 |
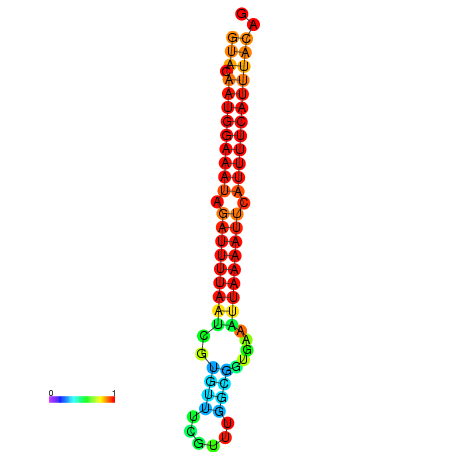
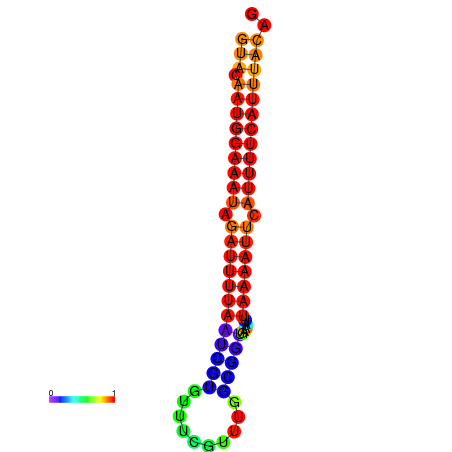
| dG=-14.0, p-value=0.009901 | dG=-13.6, p-value=0.009901 | dG=-13.5, p-value=0.009901 | dG=-13.4, p-value=0.009901 | dG=-13.0, p-value=0.009901 |
|---|---|---|---|---|
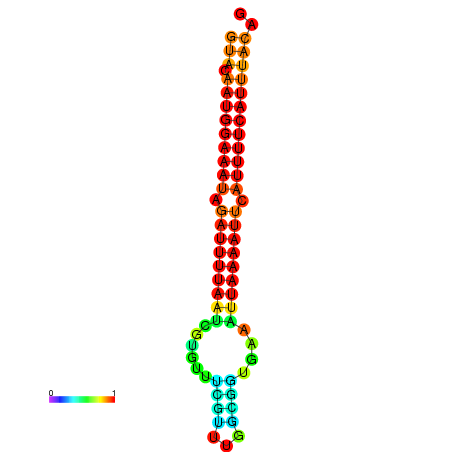 |
 |
 |
 |
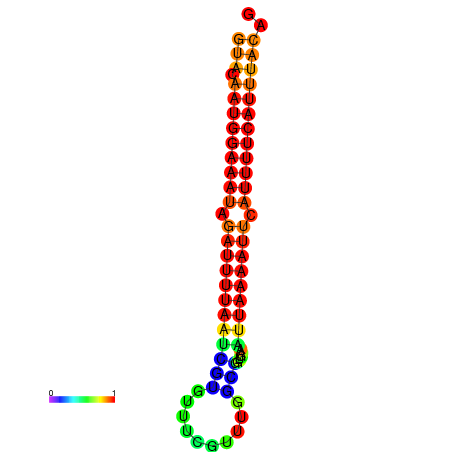 |
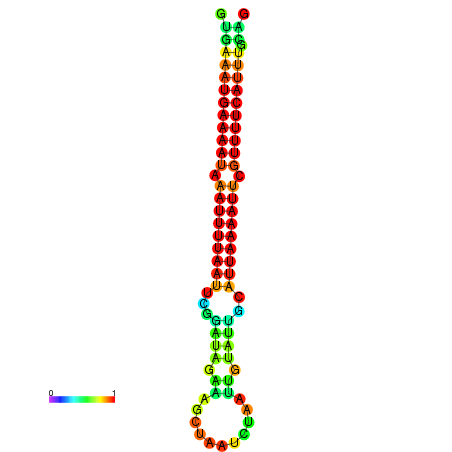
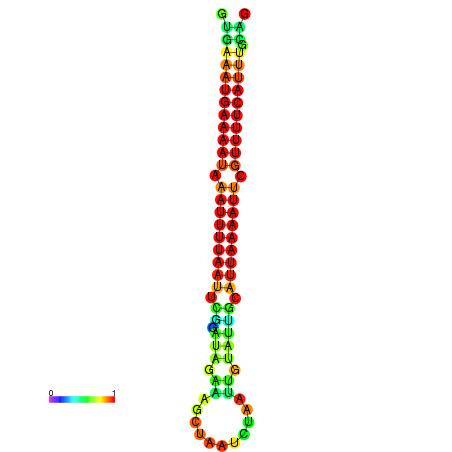
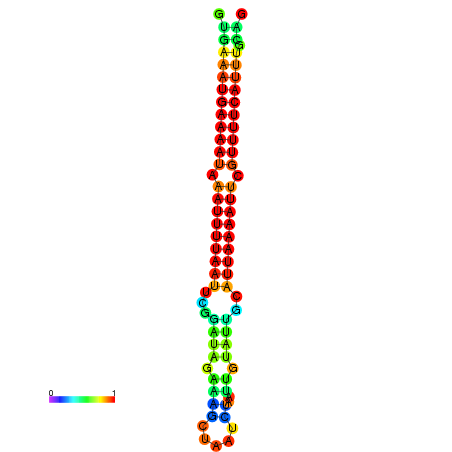
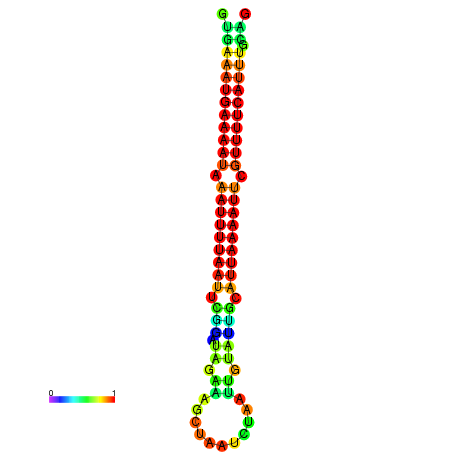
| dG=-14.7, p-value=0.009901 | dG=-14.4, p-value=0.009901 | dG=-14.1, p-value=0.009901 | dG=-14.1, p-value=0.009901 | dG=-13.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-1.7, p-value=0.009901 |
|---|
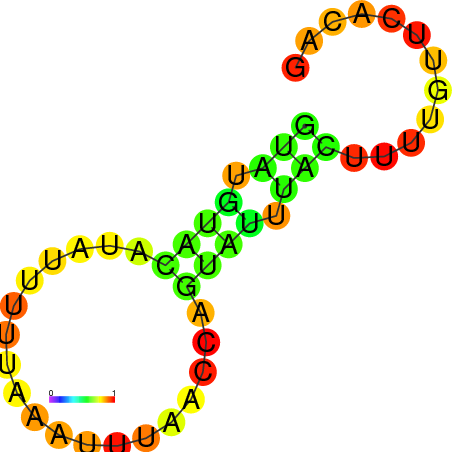 |
| dG=-7.8, p-value=0.009901 | dG=-7.8, p-value=0.009901 | dG=-7.6, p-value=0.009901 | dG=-7.6, p-value=0.009901 | dG=-7.2, p-value=0.009901 | dG=-5.9, p-value=0.009901 |
|---|---|---|---|---|---|
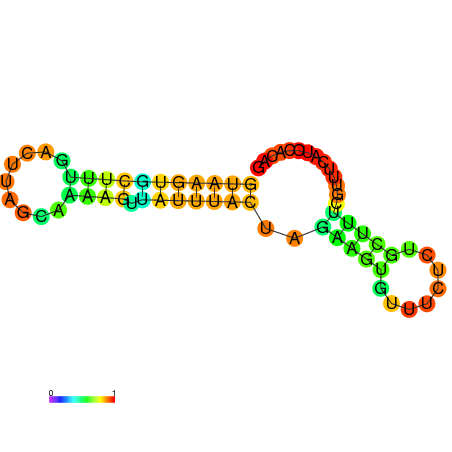 |
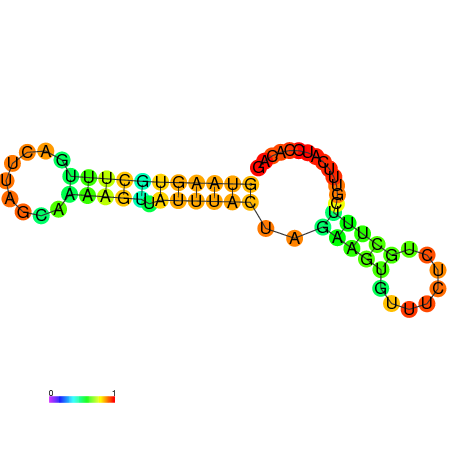 |
 |
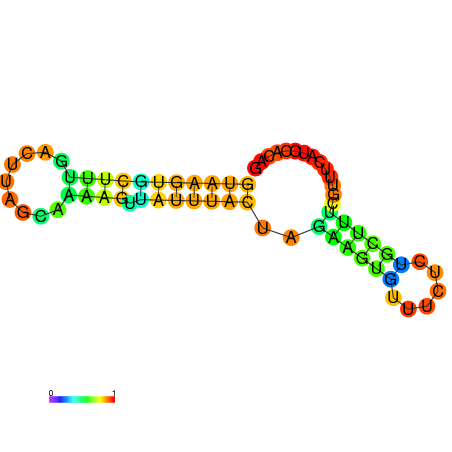 |
 |
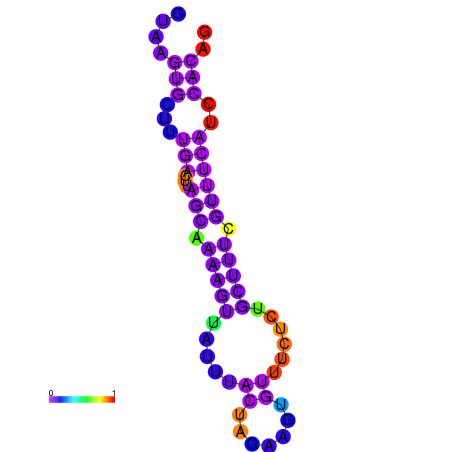 |
| dG=-7.1, p-value=0.009901 | dG=-7.0, p-value=0.009901 | dG=-7.0, p-value=0.009901 | dG=-7.0, p-value=0.009901 | dG=-6.9, p-value=0.009901 | dG=-5.3, p-value=0.009901 |
|---|---|---|---|---|---|
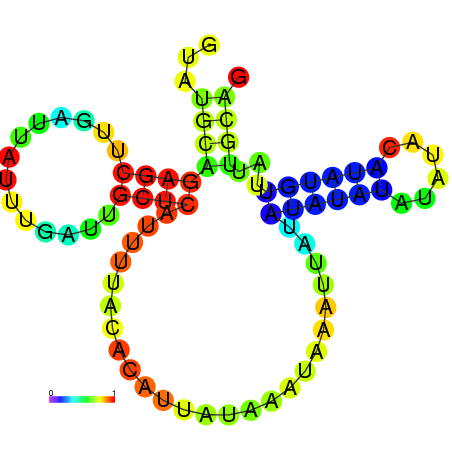 |
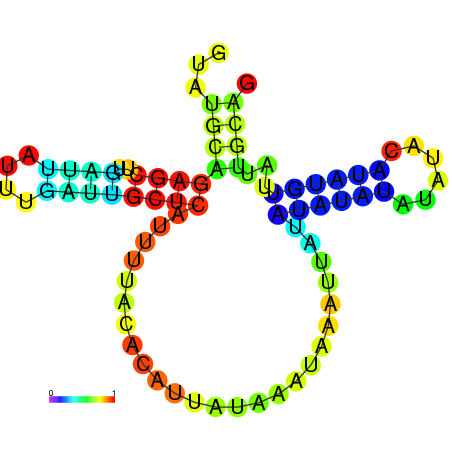 |
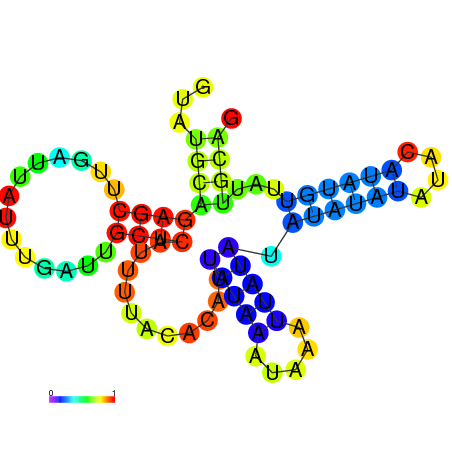 |
 |
 |
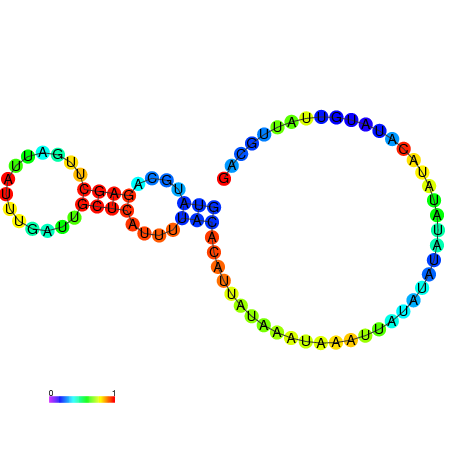 |
Generated: 03/07/2013 at 04:49 PM