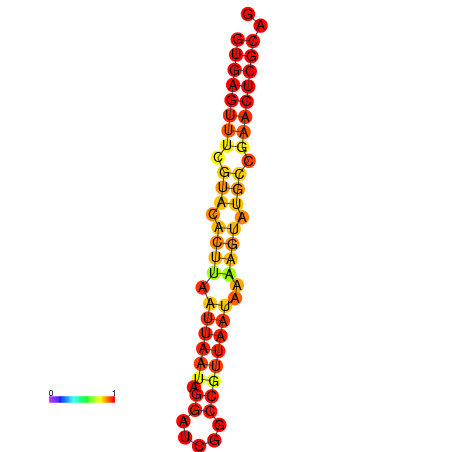
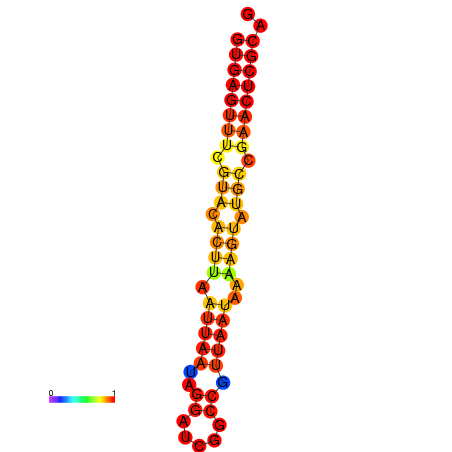
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:26617343-26617433 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAAT------------AAAAGTA-----T-GCCGAACTCGC-------AGGCGACGGATGCGCGA |
| droSim1 | chr3R:26257486-26257576 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAAT------------AAAAGTA-----T-GCCGAACTCGC-------AGGCGACGGATGCGCGA |
| droSec1 | super_4:5435200-5435290 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAAT------------AAAAGTT-----T-GCCGAACTCGC-------AGGCGACGGATGCGCGA |
| droYak2 | chr3R:27538983-27539073 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATTGGGACGGCCGTTAAT------------AAAAGTA-----T-GCCGAACTCGC-------AGGCGACGGATGCGCGA |
| droEre2 | scaffold_4820:1381504-1381594 + | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATTGGAGCGGCCGTTAAT------------AAAAGTA-----T-GCCGAACTCGC-------AGGCGACGGATGCGCGA |
| droAna3 | scaffold_12911:4366271-4366356 + | CACTGTGCTGGAGAGGTAATCTTTGAATAATTATCTGTGAGTTGTGGCATCTAAT------------GATTGT-----------TATCTTCC-------AGACGTCGCATGCGTGA |
| dp4 | chr2:577090-577184 + | CACTGTGCTGGAGCGGTAAGT----------CCATGAATTGCATCCCCCTTTGATTATTCTTTAATCTGG---AAATCCC-TGTGATCCCAT-------AGACGACGGATGCGCGA |
| droPer1 | super_7:3361274-3361368 + | CACTGTGCTGGAGCGGTAAGT----------CCATGAATTGCATCCCCCTTTGATTATTCTTTAATCTGG---AAATCCC-TGTGATCCCAT-------AGACGACGGATGCGCGA |
| droWil1 | scaffold_181130:10168431-10168520 - | AACTGTATTAGAAAGGTATAATGAATAAACCAATTTGAATAG-CAAACCATTAAA------------TCAATTT-----C-CTTTTTTGTCT-------AGACGACGTATGCGTGA |
| droVir3 | scaffold_13047:11434928-11435028 - | CACAGTGCTGGAGAGGTGGGT-GCATCCAC---------ATCAGCTGCT--TGATAACCATTTGT--AAATGTAAATCCTCCTTTAACTCG-TTTGAACAGGCGACGCATGCGCGA |
| droMoj3 | scaffold_6540:30312688-30312787 + | CACAGTGCTGGAGAGGTGGGA----------GCATTGATTTCACCACCCATAAG--ACACATTCT--CACAGTTTTTCTTGCT-TAATTCC-TTTGTACAGGCGGCGCATGCGCGA |
| droGri2 | scaffold_14624:1473562-1473665 - | CACAGTGCTGGAAAGGTGATTCAAAT-----CCATAAGCCACAATCTTCTT-AAATATTATTTTCGCTAA---TGCT--A-TTTGTACTCGCTTTGATTAGGCGTCGCATGCGGGA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:26617343-26617433 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA |
| droSim1 | chr3R:26257486-26257576 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA |
| droSec1 | super_4:5435200-5435290 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTTTGCCGAACTCGCAGGCGACGGATGCGCGA |
| droYak2 | chr3R:27538983-27539073 - | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATTGGGACGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA |
| droEre2 | scaffold_4820:1381504-1381594 + | CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATTGGAGCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-17.4, p-value=0.009901 | dG=-16.5, p-value=0.009901 |
|---|---|
 |
 |
| dG=-17.4, p-value=0.009901 | dG=-16.5, p-value=0.009901 |
|---|---|
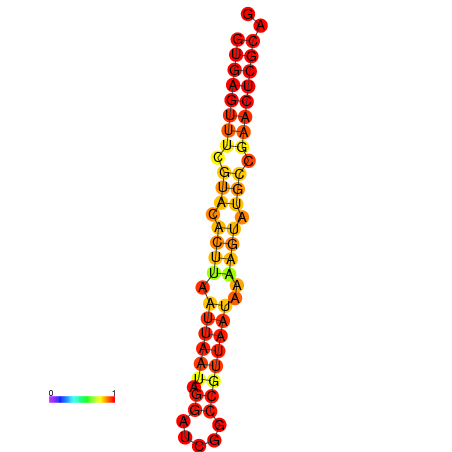 |
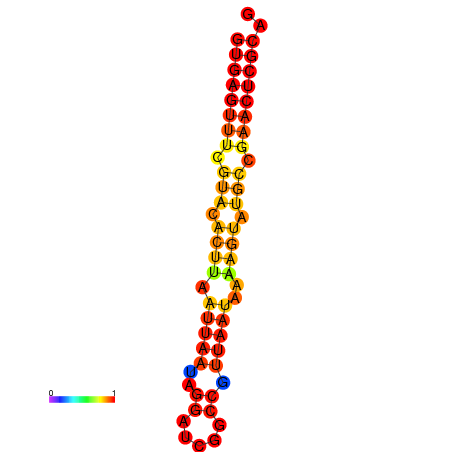 |
| dG=-17.4, p-value=0.009901 | dG=-16.5, p-value=0.009901 |
|---|---|
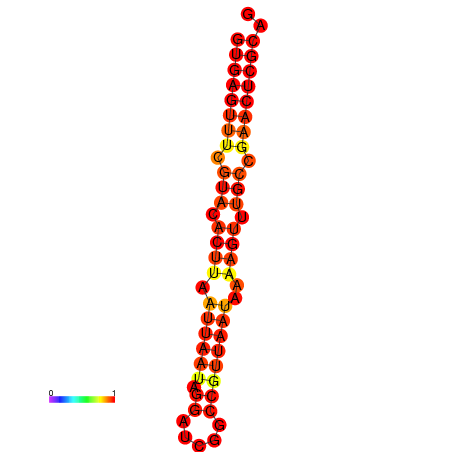 |
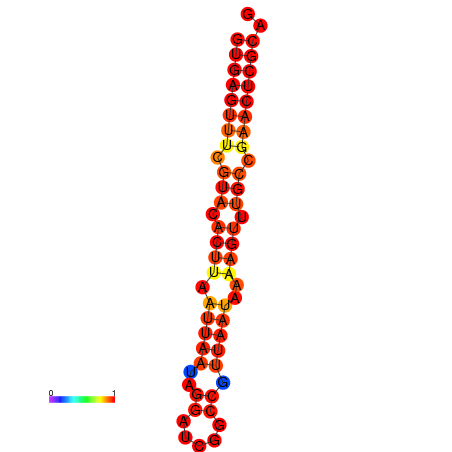 |
| dG=-18.3, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|
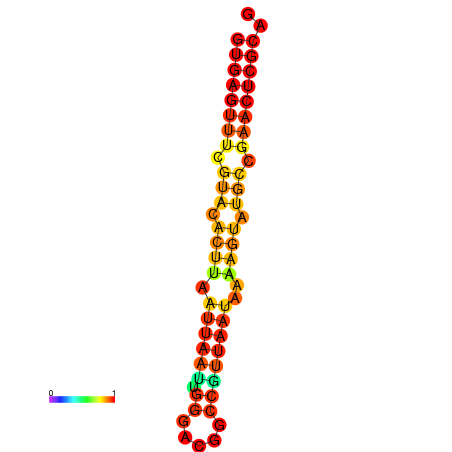 |
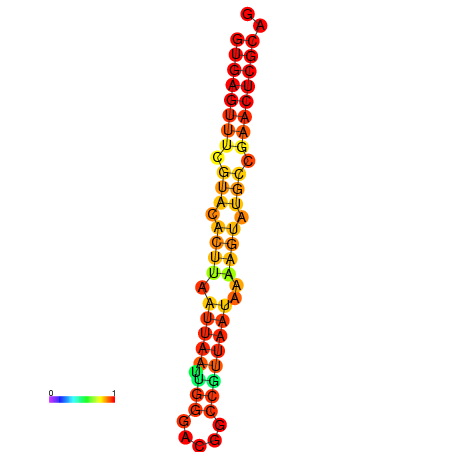 |
| dG=-17.4, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|
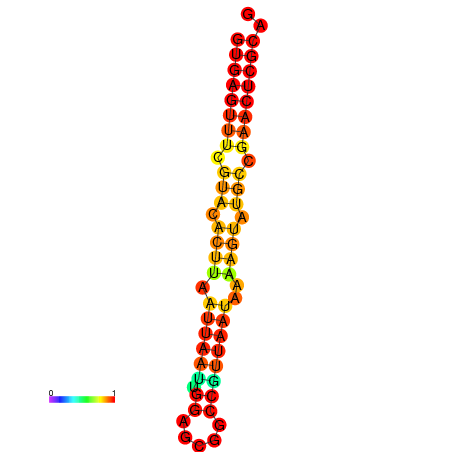 |
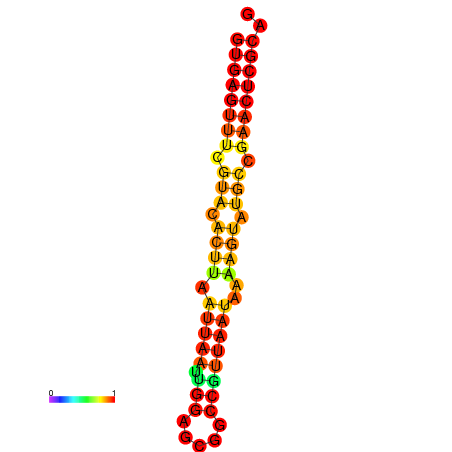 |
| dG=-6.8, p-value=0.009901 | dG=-6.8, p-value=0.009901 | dG=-6.7, p-value=0.009901 | dG=-6.7, p-value=0.009901 | dG=-5.8, p-value=0.009901 |
|---|---|---|---|---|
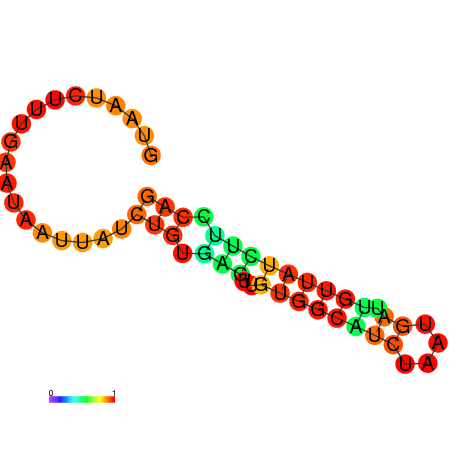 |
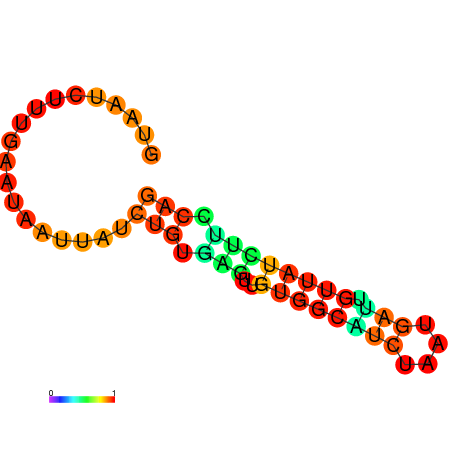 |
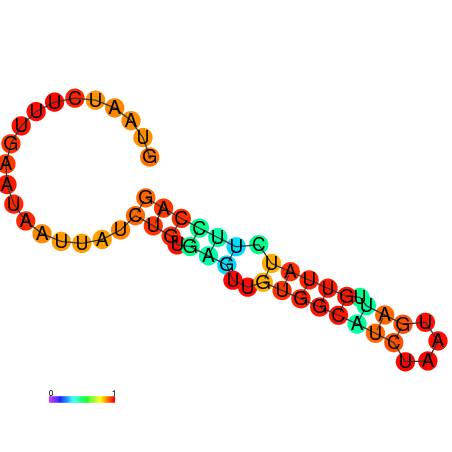 |
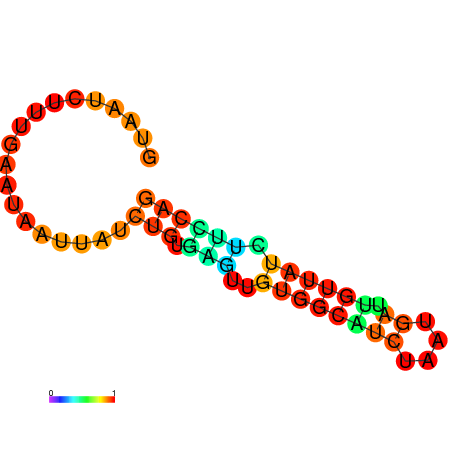 |
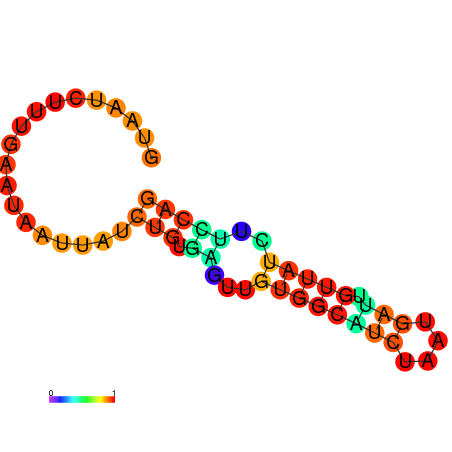 |
| dG=-6.6, p-value=0.009901 | dG=-6.4, p-value=0.009901 | dG=-6.3, p-value=0.009901 | dG=-6.0, p-value=0.009901 | dG=-5.7, p-value=0.009901 | dG=-4.5, p-value=0.009901 |
|---|---|---|---|---|---|
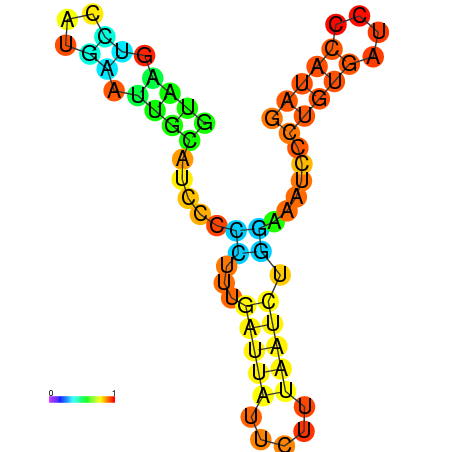 |
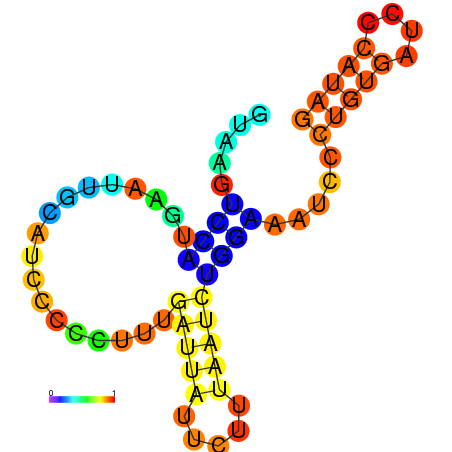 |
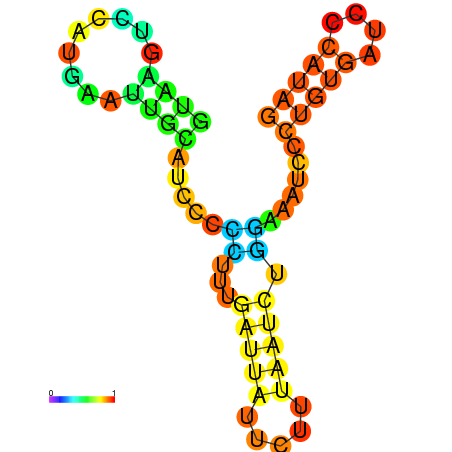 |
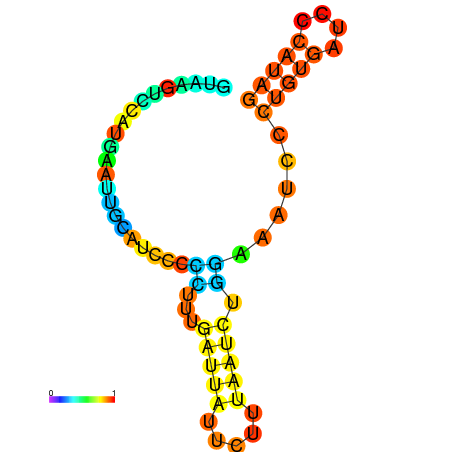 |
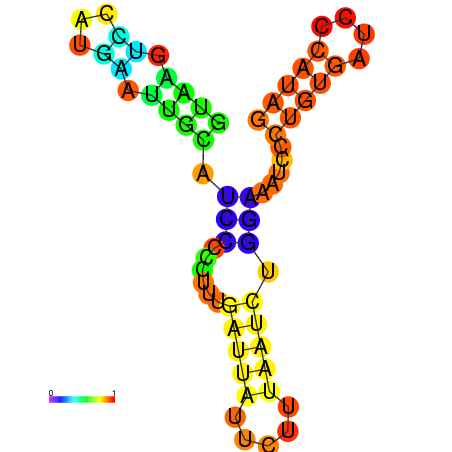 |
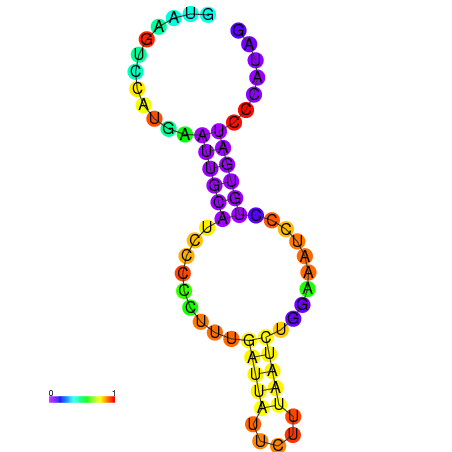 |
| dG=-6.6, p-value=0.009901 | dG=-6.4, p-value=0.009901 | dG=-6.3, p-value=0.009901 | dG=-6.0, p-value=0.009901 | dG=-5.7, p-value=0.009901 | dG=-4.5, p-value=0.009901 |
|---|---|---|---|---|---|
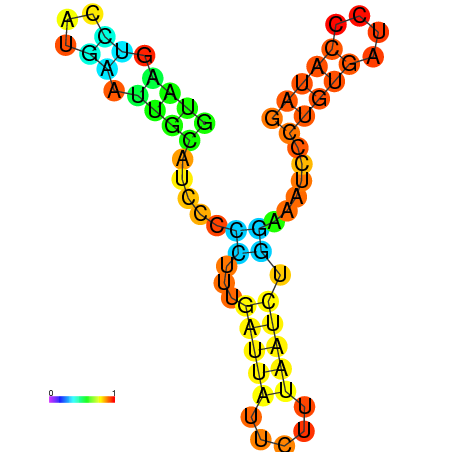 |
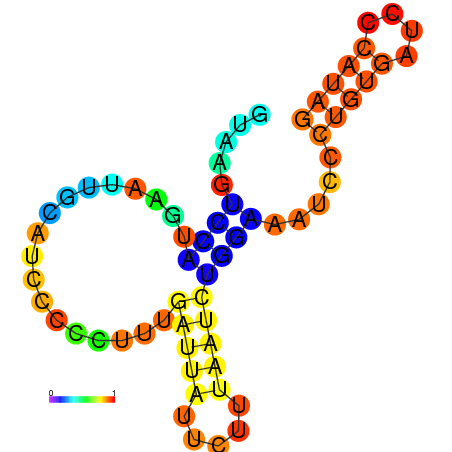 |
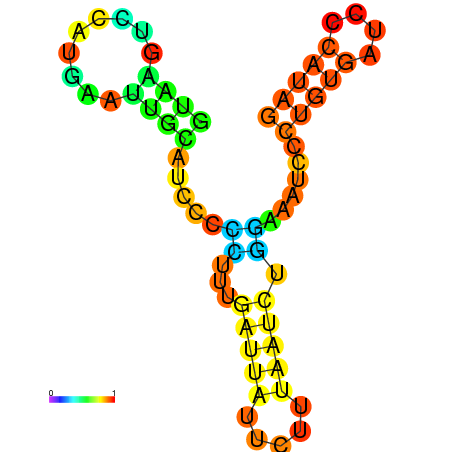 |
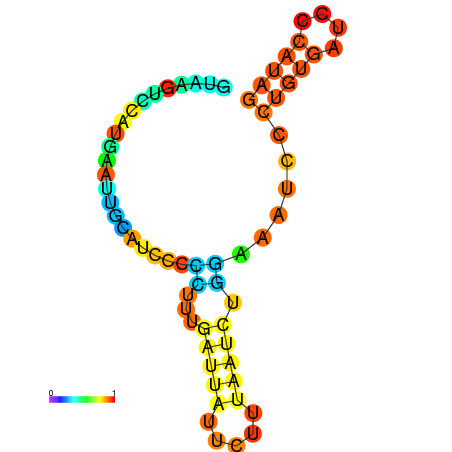 |
 |
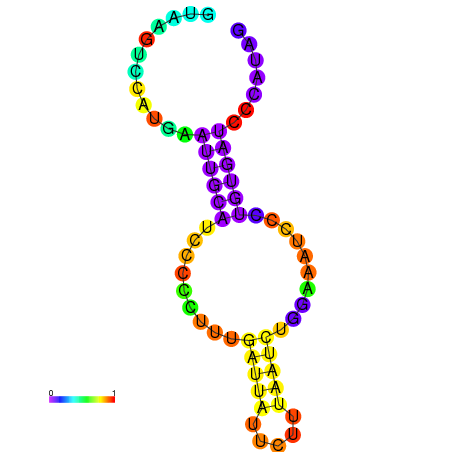 |
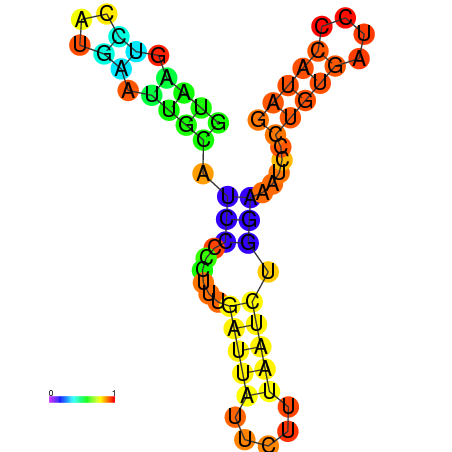
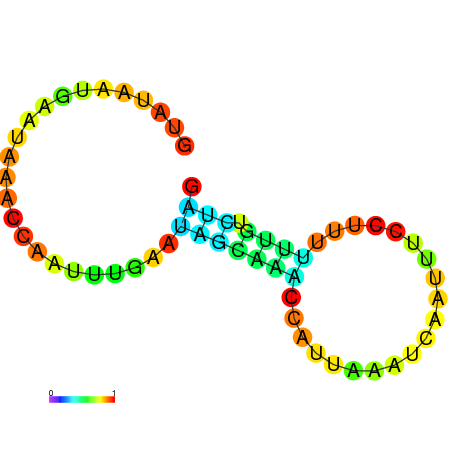
| dG=-0.5, p-value=0.019802 | dG=0.0, p-value=0.009901 | dG=0.0, p-value=0.029703 | dG=0.1, p-value=0.019802 | dG=0.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-5.8, p-value=0.009901 | dG=-5.7, p-value=0.009901 | dG=-5.7, p-value=0.009901 | dG=-5.7, p-value=0.009901 | dG=-5.6, p-value=0.009901 | dG=-4.8, p-value=0.009901 |
|---|---|---|---|---|---|
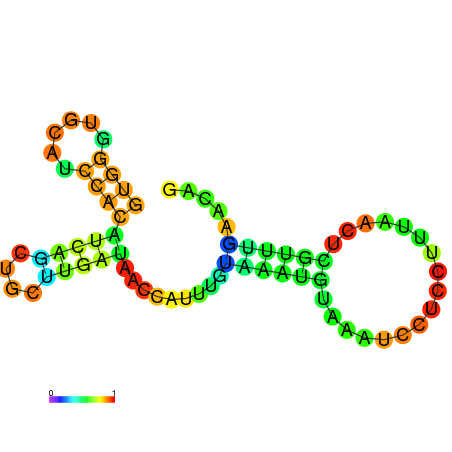 |
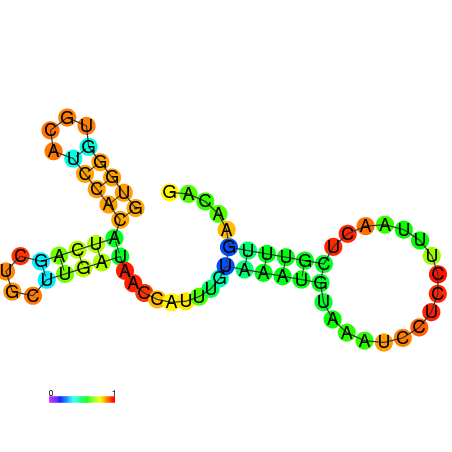 |
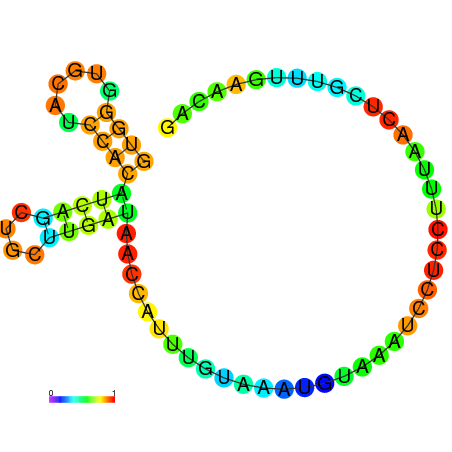 |
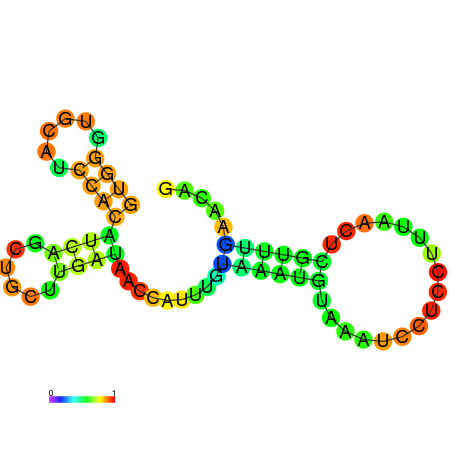 |
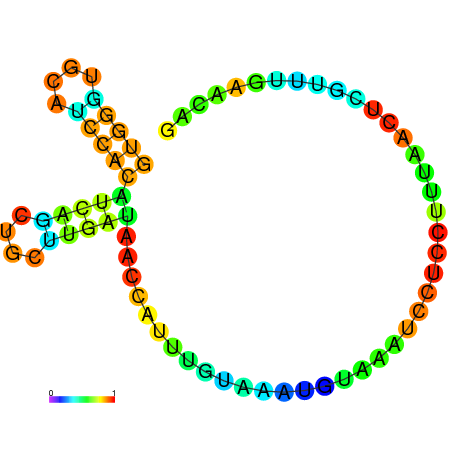 |
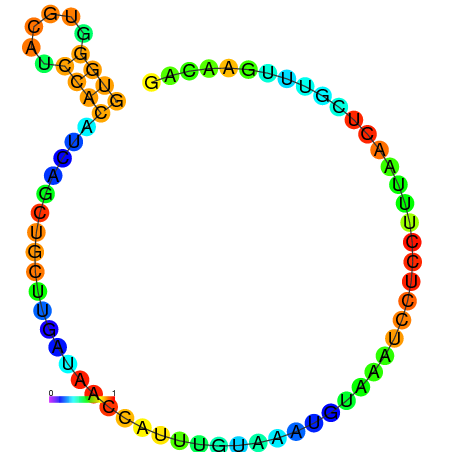 |
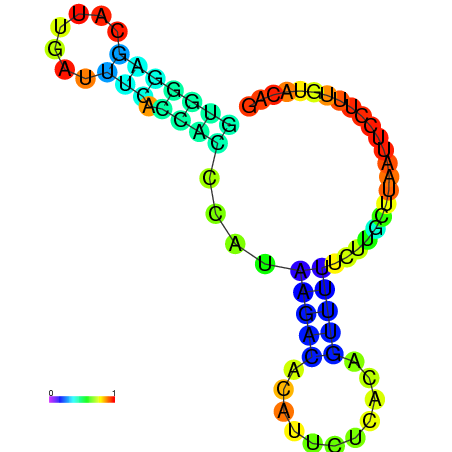
| dG=-5.0, p-value=0.009901 | dG=-4.9, p-value=0.009901 | dG=-4.7, p-value=0.009901 | dG=-4.7, p-value=0.009901 | dG=-4.6, p-value=0.009901 |
|---|---|---|---|---|
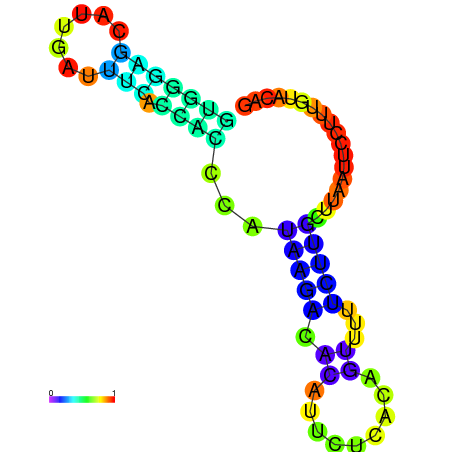 |
 |
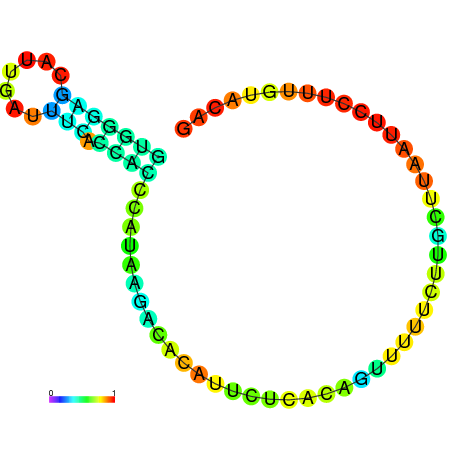 |
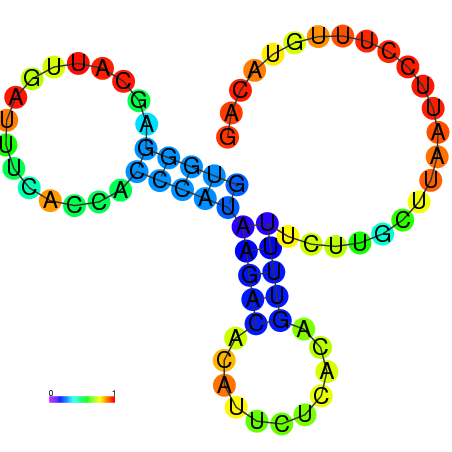 |
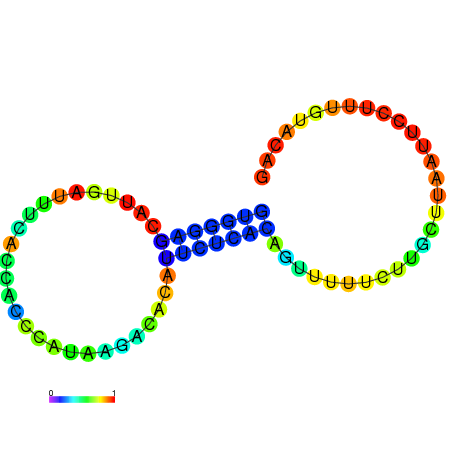 |
| dG=-6.3, p-value=0.009901 |
|---|
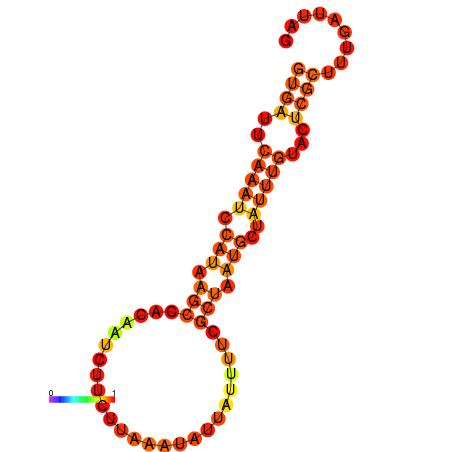 |
Generated: 03/07/2013 at 04:48 PM