| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:16679012-16679095 - | ATTCTGACCGTGCACGTGAGTTTTTGAGCCAGGAATAT--------AGTTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droSim1 | chr3R:4780974-4781057 + | ATTCTGACCGTGCACGTGAGTTTTTGAGCCAGGAATAT--------AGTTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droSec1 | super_27:54817-54900 + | ATTCTGACCGTGCACGTGAGTTTTTGAGCCAGGAATAT--------AGTTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droYak2 | chr3R:5477803-5477886 - | ATTCTGACCGTGCATGTGAGTTTTTGAGCCAGGAATAT--------AATTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droEre2 | scaffold_4770:4975828-4975911 + | ATTCTGACCGTCCACGTGAGTTTTTGAGCCATGAATAT--------AATTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droAna3 | scaffold_13340:7952070-7952154 - | GTGGTGACGGTGCATGTGAGTCTTTGAACCAGGAATAT--------AATTT----G-------TATATATTGGTTCAAATCGCTCGTAGAATCCTCCCTGGCAA |
| dp4 | chr2:12756663-12756757 + | ATTGTGACGATGCATGTGAGATTTTGAATCTAATATAT--------AATATAATCCGTACGTGTAT-ATATGGTTCAAATTACTCGTAGAACCCTCCCTGGCAA |
| droPer1 | super_0:7702112-7702205 - | ATTGTGACGATGCATGTGAGATTTTGAATCTAATATAT--------AATATAATCT-TACATGTAT-ATATGGTTCAAATAACTCGTAGAATCCTCCCTGGCAA |
| droWil1 | scaffold_181108:2184466-2184552 + | ATTCTGACCGTACATGTGAGTCTTTGAATCAGGAATAT--------ATTATCA--T-------TATATATTGGTTCGAAATGCTCGTAGAATCCACCTTGGCAA |
| droVir3 | scaffold_13047:12031331-12031423 + | ATTGTCACGGTGCATGTGAGTCATTGAACCAGGAATATATGTATGTAATTC----T-------TATATATTGGTTCAAATTTCTCGCAGAATCCGCCCTGGCAA |
| droMoj3 | scaffold_6540:29750589-29750676 - | ATTGTGACAGTGCATGTGAGTCTTTGAGCCAGGAATAT--------ATGTTCATCT-------TAT-TATTGGTTCAAATCTCTCGTAGAATCCTCCCTGGCAA |
| droGri2 | scaffold_14830:1201859-1201943 + | ATTGTCACTGAGCATGTGAGTCTTTGAGCCAGGAATAT--------AATTG----T-------TATATATTGGTTCAAACAACTCGTAGAATCCGCCCTGGCAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:16679012-16679095 - | ATTCTGACCGTGCACGTGAGTTTTTGAGCCAGGAATAT--------AGTTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droSim1 | chr3R:4780974-4781057 + | ATTCTGACCGTGCACGTGAGTTTTTGAGCCAGGAATAT--------AGTTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droSec1 | super_27:54817-54900 + | ATTCTGACCGTGCACGTGAGTTTTTGAGCCAGGAATAT--------AGTTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droYak2 | chr3R:5477803-5477886 - | ATTCTGACCGTGCATGTGAGTTTTTGAGCCAGGAATAT--------AATTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droEre2 | scaffold_4770:4975828-4975911 + | ATTCTGACCGTCCACGTGAGTTTTTGAGCCATGAATAT--------AATTC----T-------TAT-TATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAA |
| droAna3 | scaffold_13340:7952070-7952154 - | GTGGTGACGGTGCATGTGAGTCTTTGAACCAGGAATAT--------AATTT----G-------TATATATTGGTTCAAATCGCTCGTAGAATCCTCCCTGGCAA |
| dp4 | chr2:12756663-12756757 + | ATTGTGACGATGCATGTGAGATTTTGAATCTAATATAT--------AATATAATCCGTACGTGTAT-ATATGGTTCAAATTACTCGTAGAACCCTCCCTGGCAA |
| droPer1 | super_0:7702112-7702205 - | ATTGTGACGATGCATGTGAGATTTTGAATCTAATATAT--------AATATAATCT-TACATGTAT-ATATGGTTCAAATAACTCGTAGAATCCTCCCTGGCAA |
| droWil1 | scaffold_181108:2184466-2184552 + | ATTCTGACCGTACATGTGAGTCTTTGAATCAGGAATAT--------ATTATCA--T-------TATATATTGGTTCGAAATGCTCGTAGAATCCACCTTGGCAA |
| droVir3 | scaffold_13047:12031331-12031423 + | ATTGTCACGGTGCATGTGAGTCATTGAACCAGGAATATATGTATGTAATTC----T-------TATATATTGGTTCAAATTTCTCGCAGAATCCGCCCTGGCAA |
| droMoj3 | scaffold_6540:29750589-29750676 - | ATTGTGACAGTGCATGTGAGTCTTTGAGCCAGGAATAT--------ATGTTCATCT-------TAT-TATTGGTTCAAATCTCTCGTAGAATCCTCCCTGGCAA |
| droGri2 | scaffold_14830:1201859-1201943 + | ATTGTCACTGAGCATGTGAGTCTTTGAGCCAGGAATAT--------AATTG----T-------TATATATTGGTTCAAACAACTCGTAGAATCCGCCCTGGCAA |
| Species | Read pileup | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||
| dp4 |
|
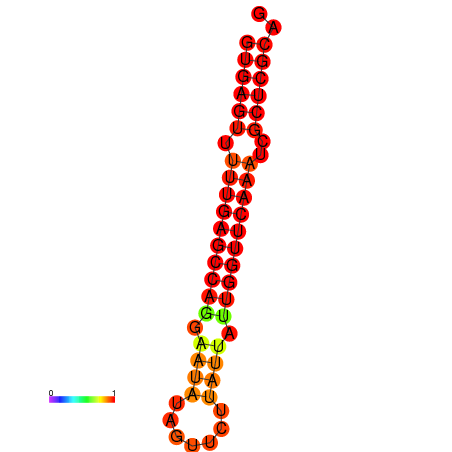
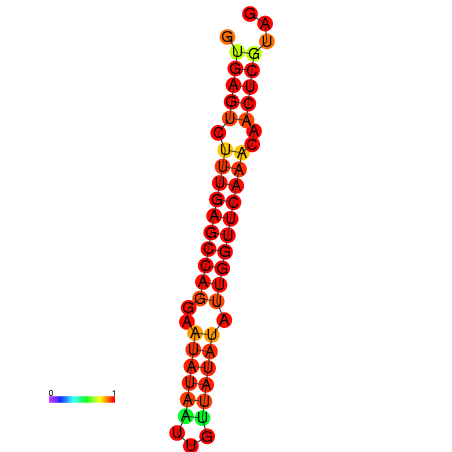
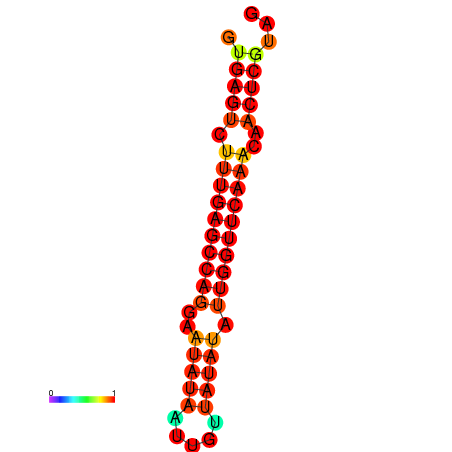
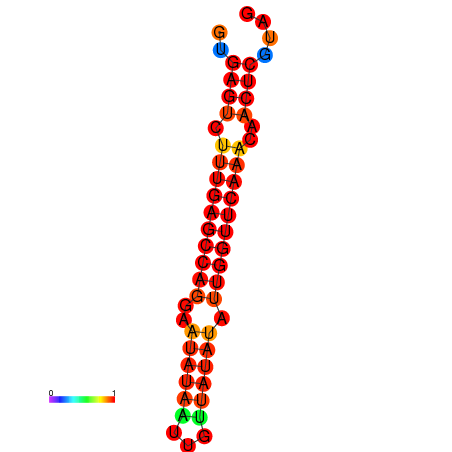
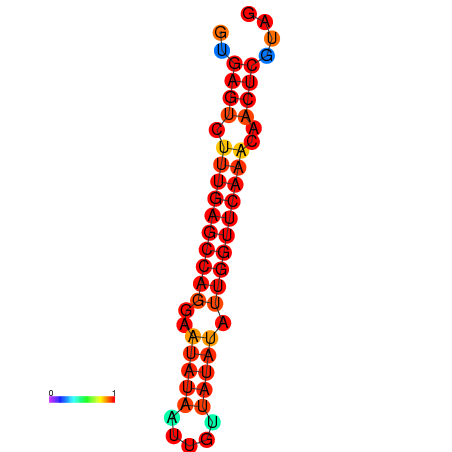
| dG=-21.0, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|
 |
 |
 |
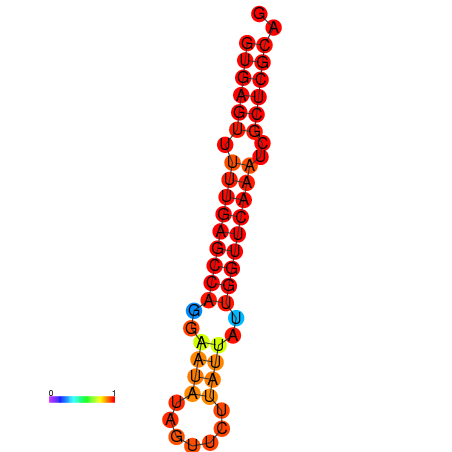
| dG=-21.0, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|
 |
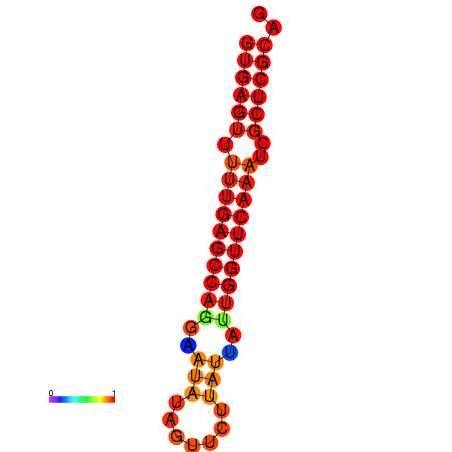
 |
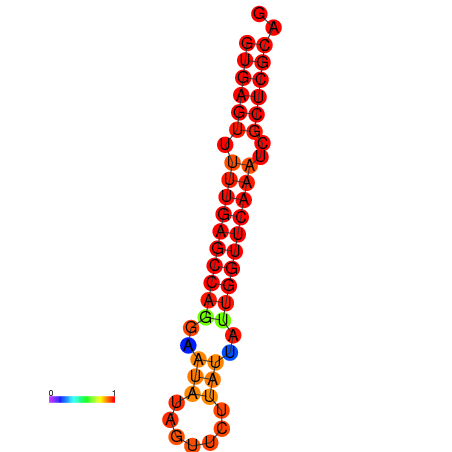 |
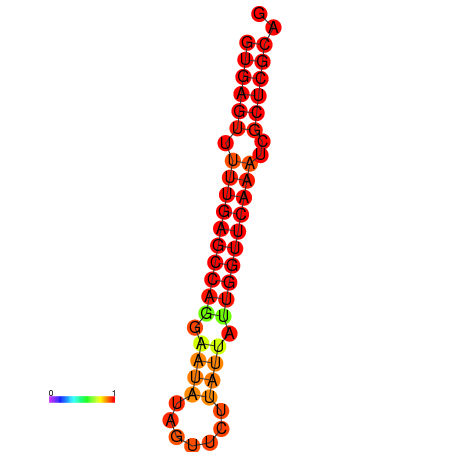
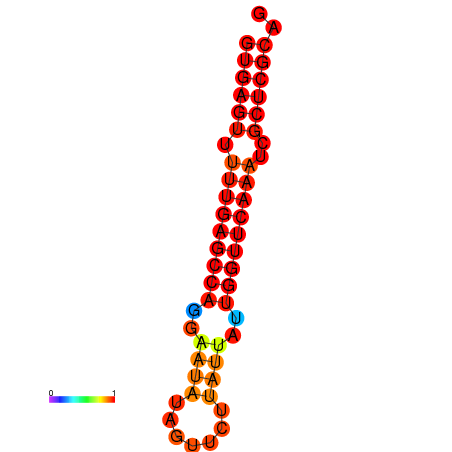
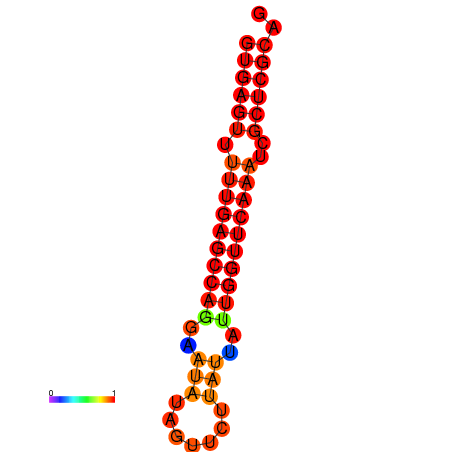
| dG=-21.0, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|
 |
 |
 |
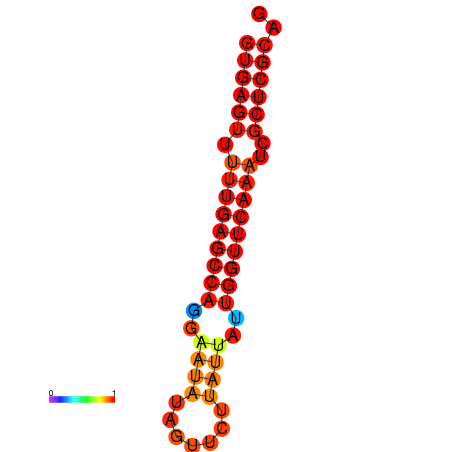
| dG=-21.0, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|
 |
 |
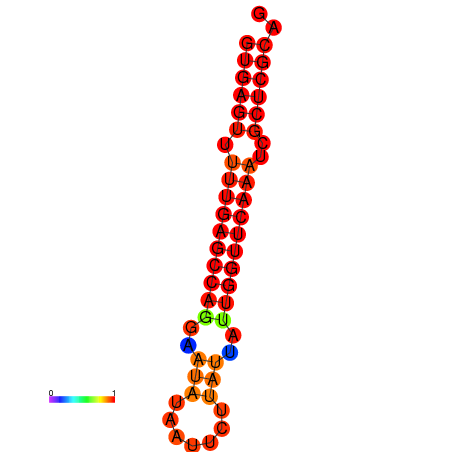 |
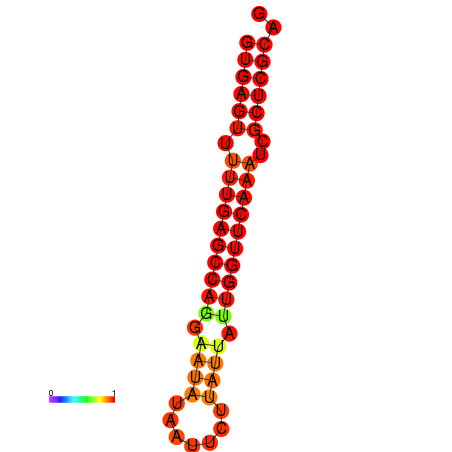
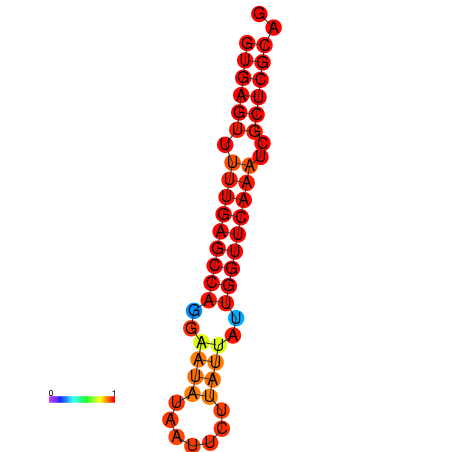
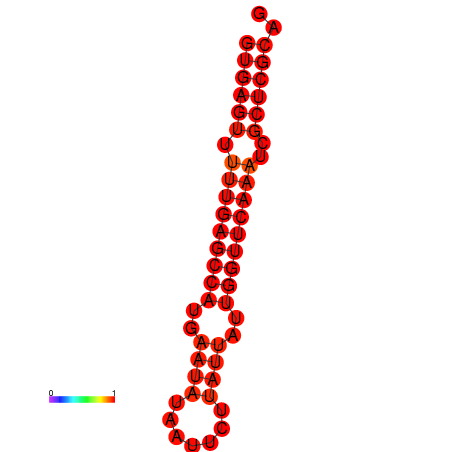
| dG=-21.6, p-value=0.009901 |
|---|
 |
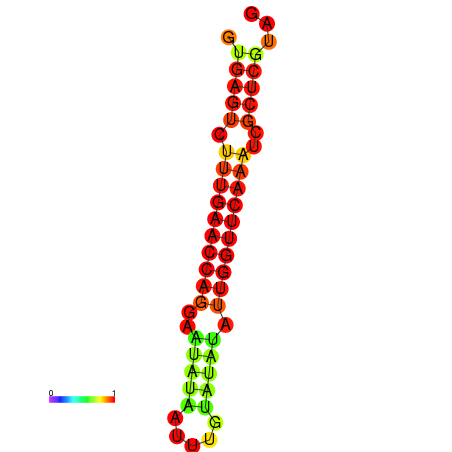
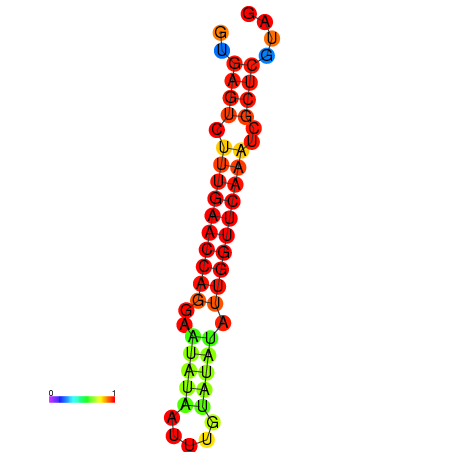
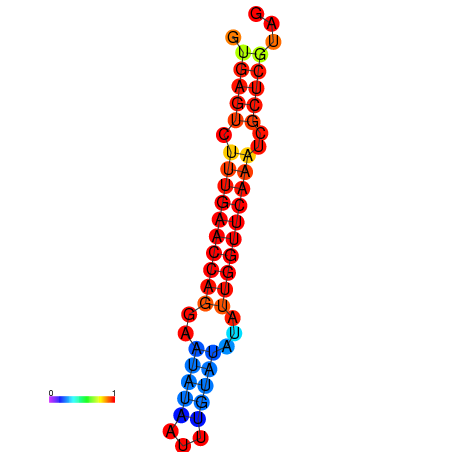
| dG=-17.2, p-value=0.009901 | dG=-16.6, p-value=0.009901 | dG=-16.4, p-value=0.009901 |
|---|---|---|
 |
 |
 |
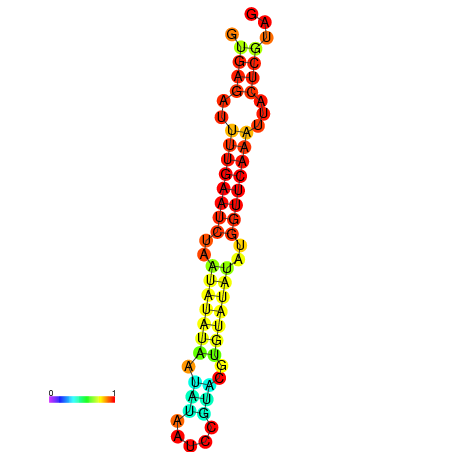
| dG=-12.3, p-value=0.009901 | dG=-12.1, p-value=0.009901 | dG=-11.7, p-value=0.009901 | dG=-11.5, p-value=0.009901 |
|---|---|---|---|
 |
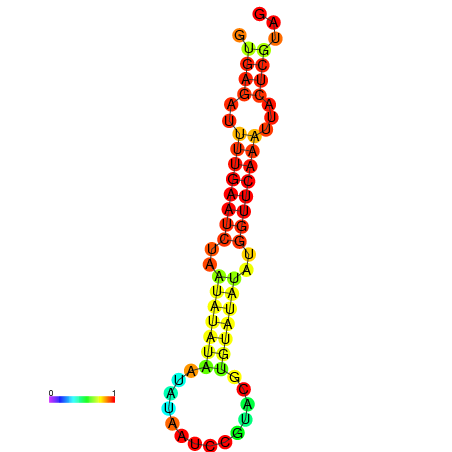 |
 |
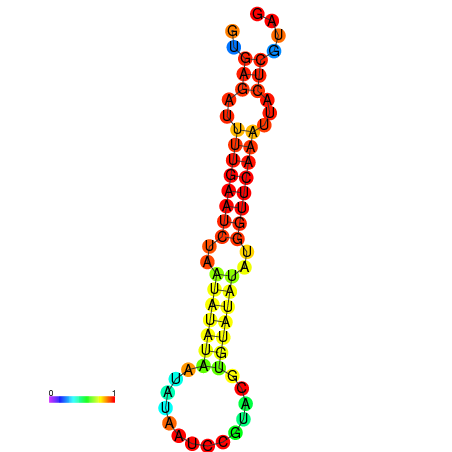 |
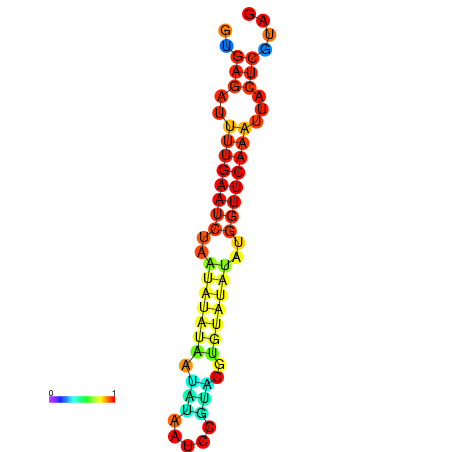
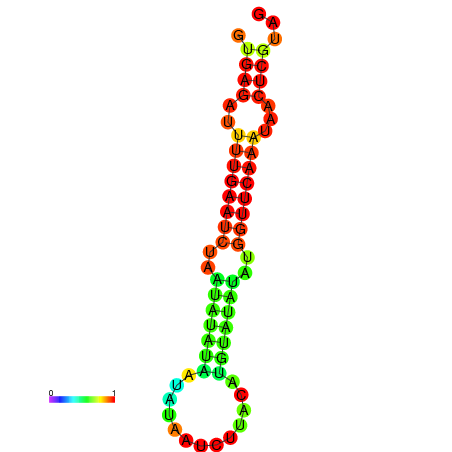
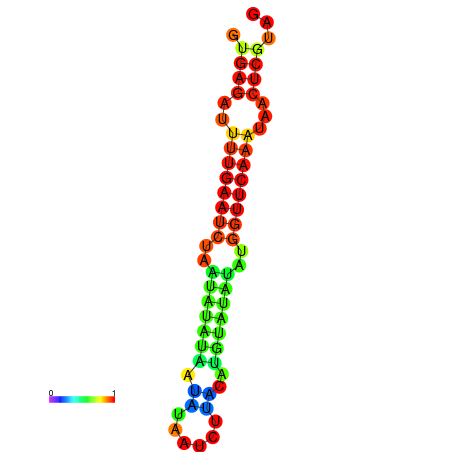
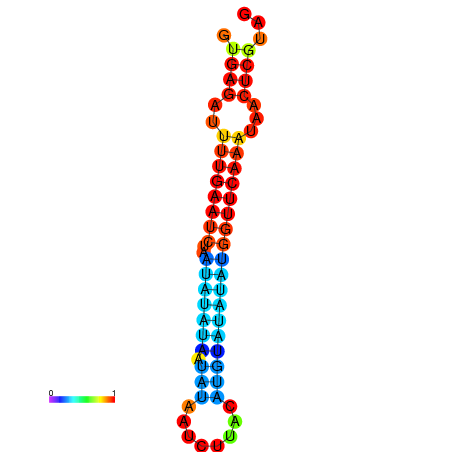
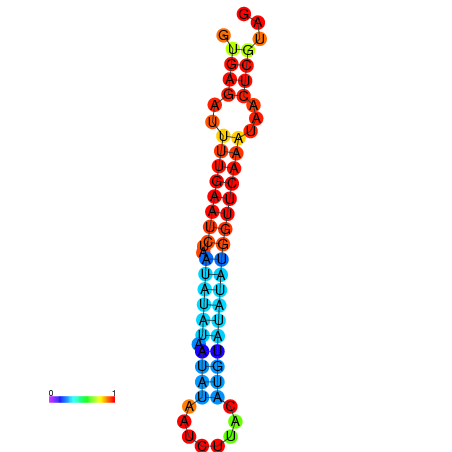
| dG=-12.2, p-value=0.009901 | dG=-12.1, p-value=0.009901 | dG=-11.7, p-value=0.009901 | dG=-11.7, p-value=0.009901 | dG=-11.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
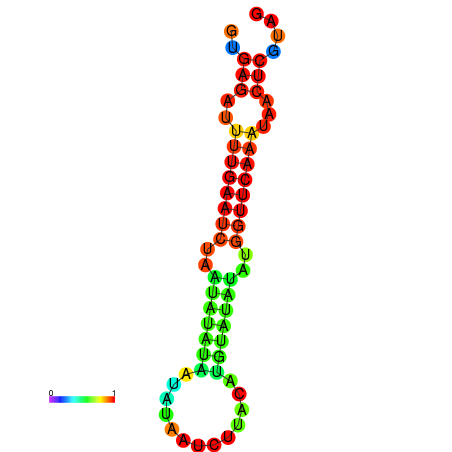 |
| dG=-14.9, p-value=0.009901 | dG=-14.3, p-value=0.009901 |
|---|---|
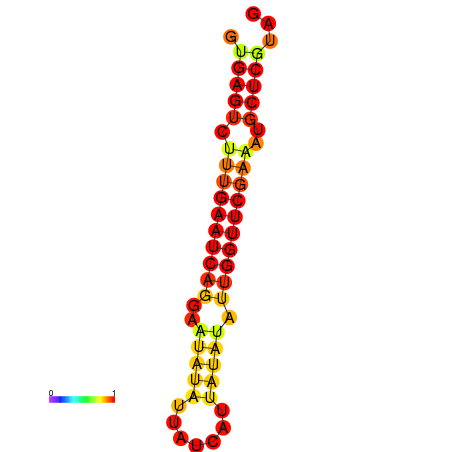 |
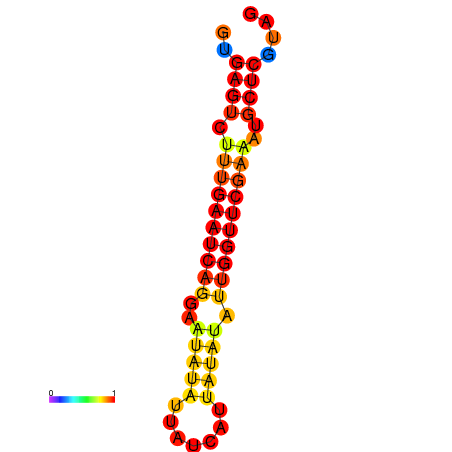 |
| dG=-17.8, p-value=0.009901 | dG=-17.7, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.3, p-value=0.009901 | dG=-17.2, p-value=0.009901 |
|---|---|---|---|---|
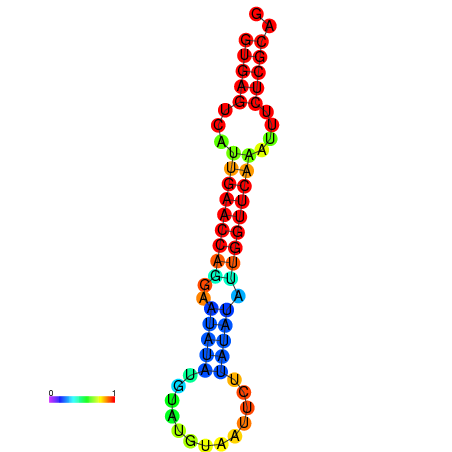 |
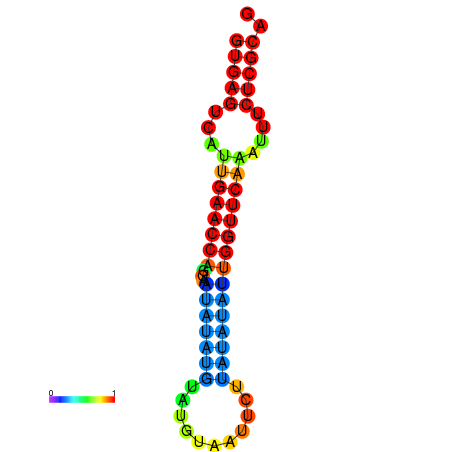 |
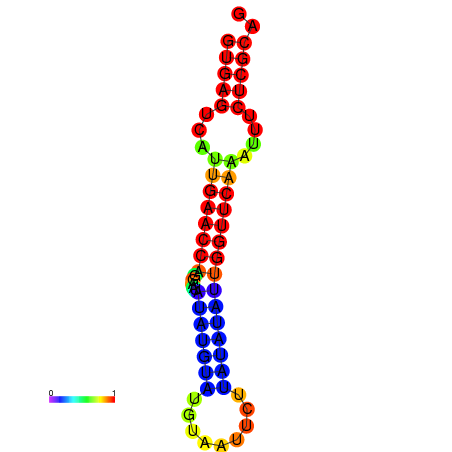 |
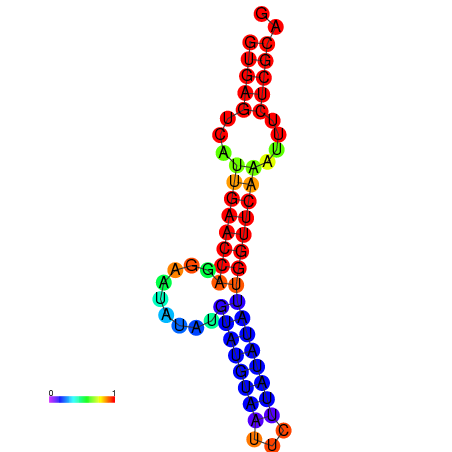 |
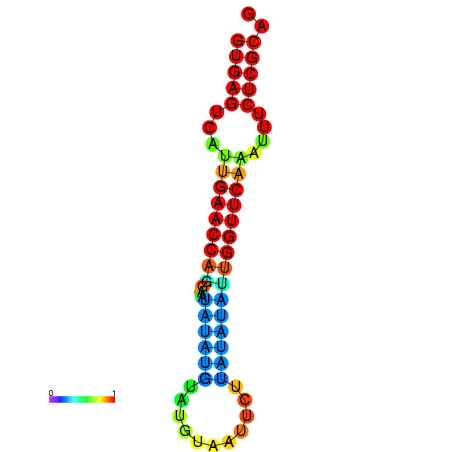 |
| dG=-14.6, p-value=0.009901 | dG=-14.4, p-value=0.009901 | dG=-14.2, p-value=0.009901 | dG=-14.0, p-value=0.009901 | dG=-14.0, p-value=0.009901 |
|---|---|---|---|---|
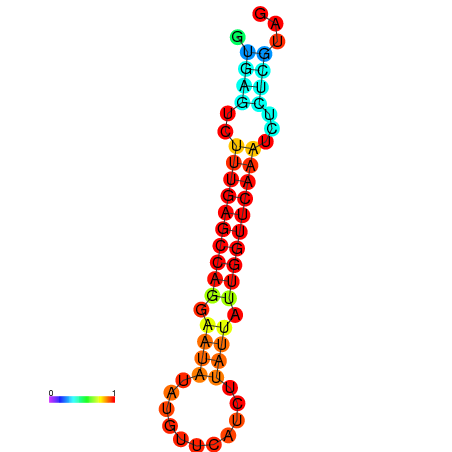 |
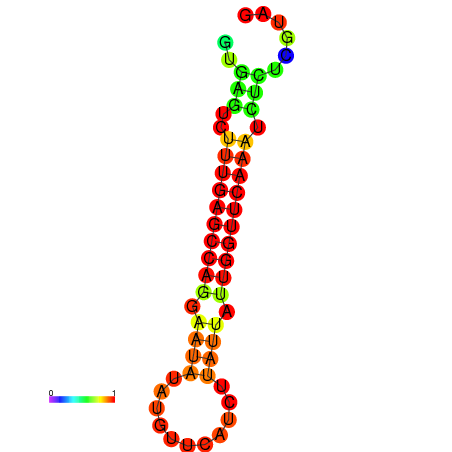 |
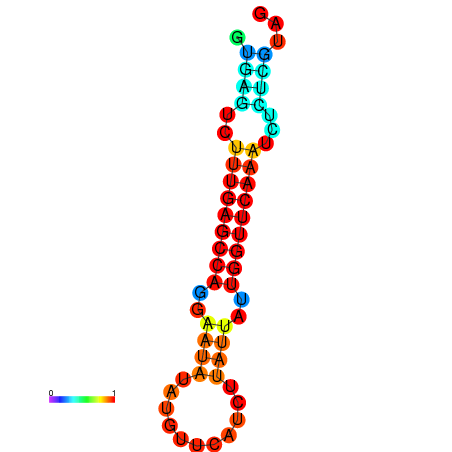 |
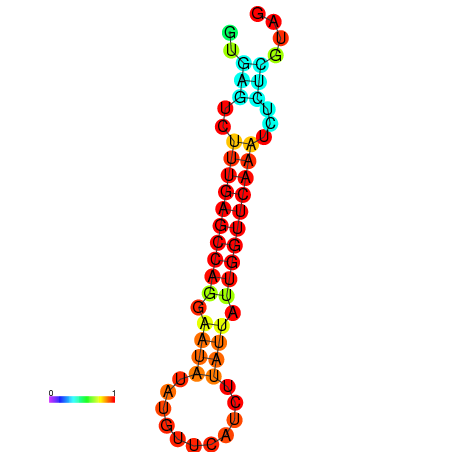 |
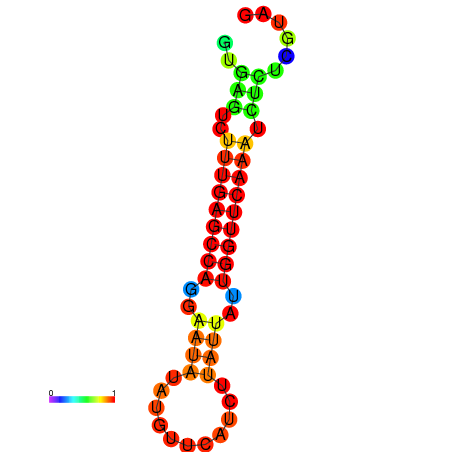 |
| dG=-17.3, p-value=0.009901 | dG=-17.1, p-value=0.009901 | dG=-16.7, p-value=0.009901 | dG=-16.5, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
Generated: 03/07/2013 at 04:47 PM