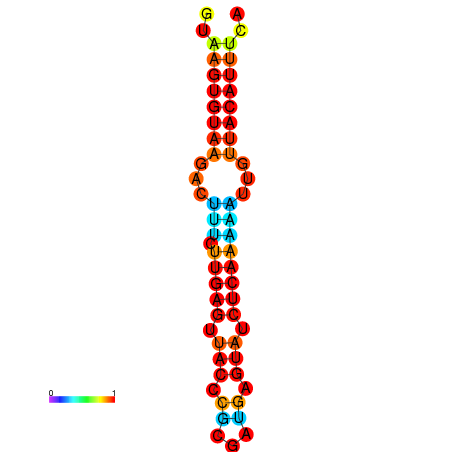
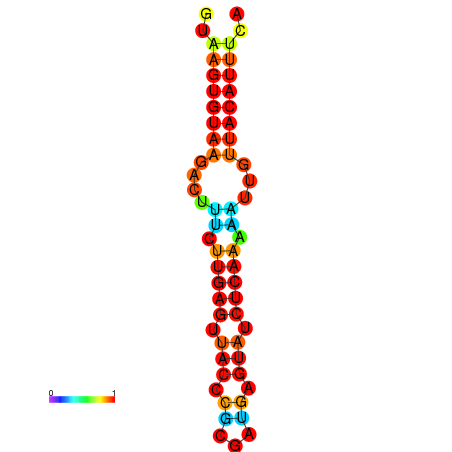

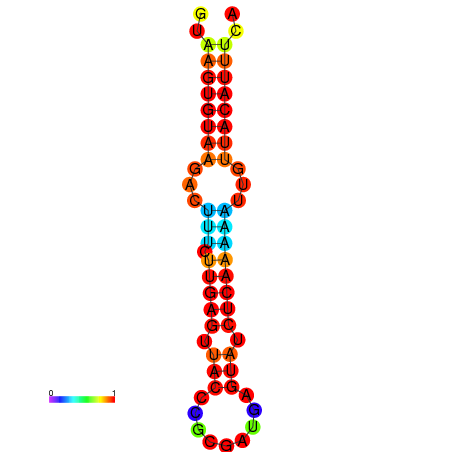
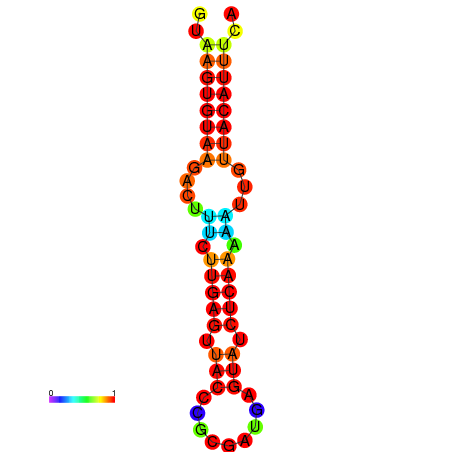
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:19880562-19880652 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTA---------CCCGCGATGAGTATCTCAAAA--ATTG-----TTACATTTCAGAGCGATAGCTATCCA |
| droSim1 | chr2R:18425639-18425729 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTA---------CCCGCGATGAGTATCTCAAAA--ATTG-----TTACATTTCAGAGCGATAGCTATCCA |
| droSec1 | super_9:3164536-3164626 - | CTCTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTA---------CCCGCGATGAGTATCTCAAAA--ATTG-----TTACATTTCAGAGCGATAGCTATCCA |
| droYak2 | chr2R:19858946-19859036 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTA---------CCCGCGAGGAGTATCTCAAAA--ATTA-----TTACATTTCAGGGCGATAGTTACCCG |
| droEre2 | scaffold_4845:21253645-21253735 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTGTTGAGTTA---------CCCACGTAGAGTATCTCAAAA--TTTG-----TTACATTTCAGAGCGATAGCTACCCA |
| droAna3 | scaffold_13266:5750531-5750620 - | CTATCGATTCAAGATGTAAGTTTTGAATACTTCTACTCTA---------TCTTGGA-ATGTCTCTCATAA--TTGT-----CCATCTTCTAGAACGATAGCTATCCA |
| dp4 | chr3:7562600-7562686 - | ATGTCCATACAAGATGTAAGTTCGAGAATC--------------CCAAACACATAAGTTCTTTTTTAACA------TCCAAATATTTTGTAGAGCGACAACATTCCA |
| droPer1 | super_2:7758003-7758089 - | ATGTCCATACAAGATGTAAGTTCGAGAATC--------------CCAAAAACATAAGTTCTTTTTTAACA------TCCAAATATTTTGTAGAGCGACAACATTCCA |
| droWil1 | scaffold_180700:5103983-5104075 + | TTATCTATCCAGGATGTAAGTATTTAAA--------GTTATTTTCAT----AATGACGGGAAATCAATGATTTGCATTC--ACTCATTGCAGAGTGATAACCTCCCA |
| droVir3 | Unknown | ----------------------------------------------------------------------------------------------------------- |
| droMoj3 | scaffold_6496:14788216-14788309 + | TTATCTCTACAGGAGGTAAGGACCAATTGGTA--AAGT------GAGAAATTGGAAGAAATTTATTGAAA--TTTA---TACTCGTGTTTAGGACAAGGAGTTCACA |
| droGri2 | Unknown | ----------------------------------------------------------------------------------------------------------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:19880562-19880652 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTACCCGCGATGAGTATCTCAAAAATTGTTACATTTCAGAGCGATAGCTATCCA |
| droSim1 | chr2R:18425639-18425729 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTACCCGCGATGAGTATCTCAAAAATTGTTACATTTCAGAGCGATAGCTATCCA |
| droSec1 | super_9:3164536-3164626 - | CTCTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTACCCGCGATGAGTATCTCAAAAATTGTTACATTTCAGAGCGATAGCTATCCA |
| droYak2 | chr2R:19858946-19859036 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTCTTGAGTTACCCGCGAGGAGTATCTCAAAAATTATTACATTTCAGGGCGATAGTTACCCG |
| droEre2 | scaffold_4845:21253645-21253735 - | CTGTCCATTCAAGACGTAAGTGTAAGACTTTGTTGAGTTACCCACGTAGAGTATCTCAAAATTTGTTACATTTCAGAGCGATAGCTACCCA |
| Species | Read pileup | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-11.6, p-value=0.009901 | dG=-11.6, p-value=0.009901 | dG=-11.3, p-value=0.009901 | dG=-11.0, p-value=0.009901 | dG=-11.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
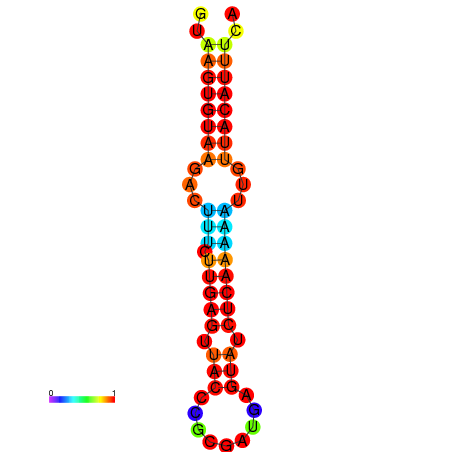 |
 |
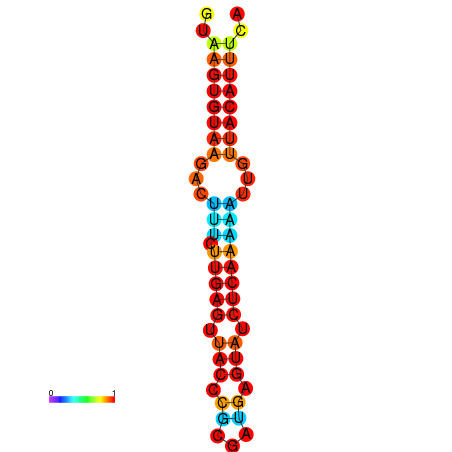
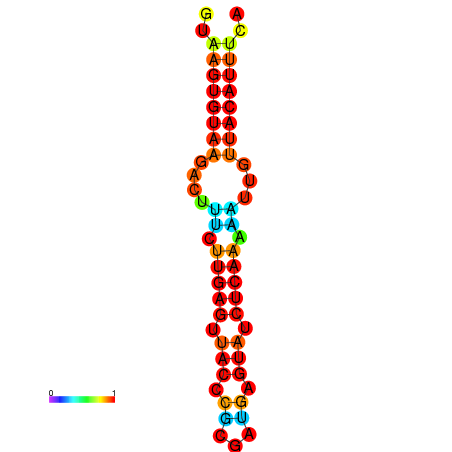
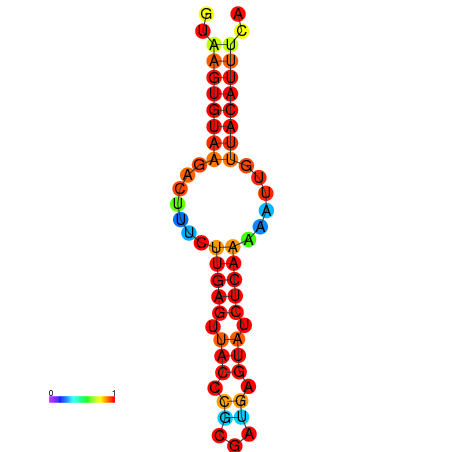
| dG=-11.6, p-value=0.009901 | dG=-11.6, p-value=0.009901 | dG=-11.3, p-value=0.009901 | dG=-11.0, p-value=0.009901 | dG=-11.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
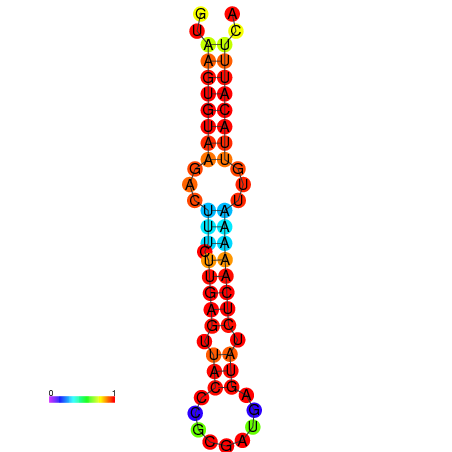 |
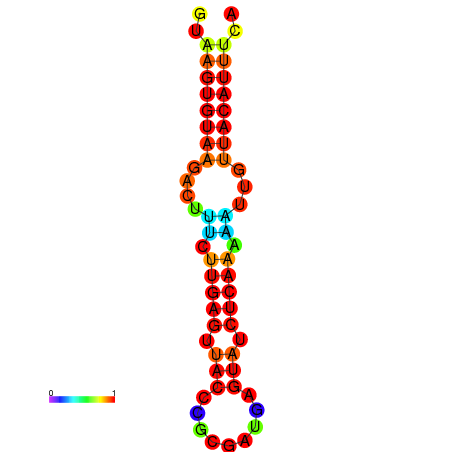 |
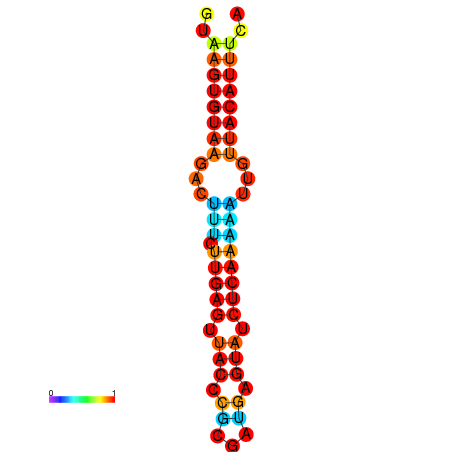
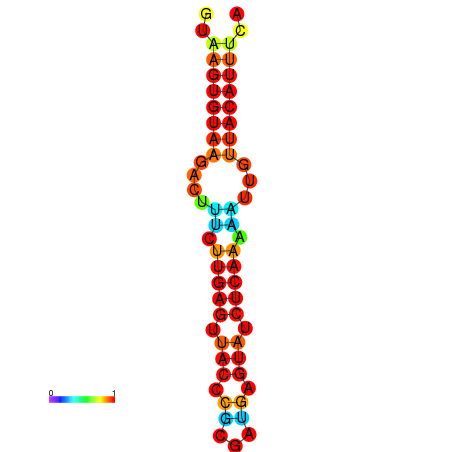
| dG=-11.6, p-value=0.009901 | dG=-11.6, p-value=0.009901 | dG=-11.3, p-value=0.009901 | dG=-11.0, p-value=0.009901 | dG=-11.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
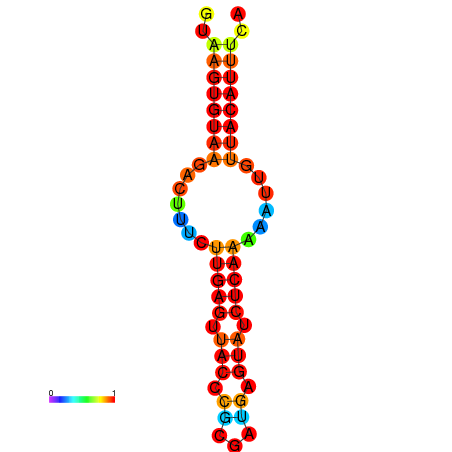 |
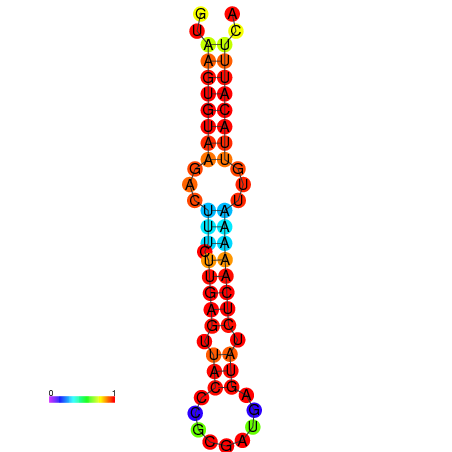 |
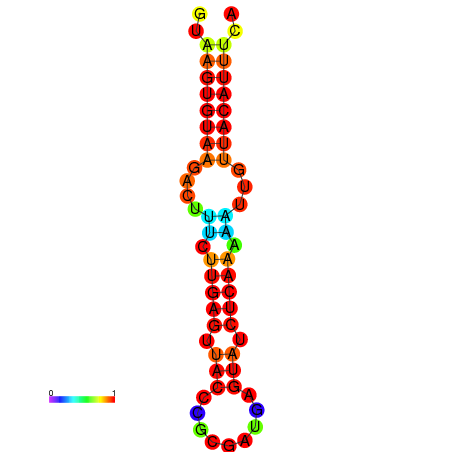 |
| dG=-15.9, p-value=0.009901 | dG=-15.9, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.2, p-value=0.009901 | dG=-15.2, p-value=0.009901 |
|---|---|---|---|---|
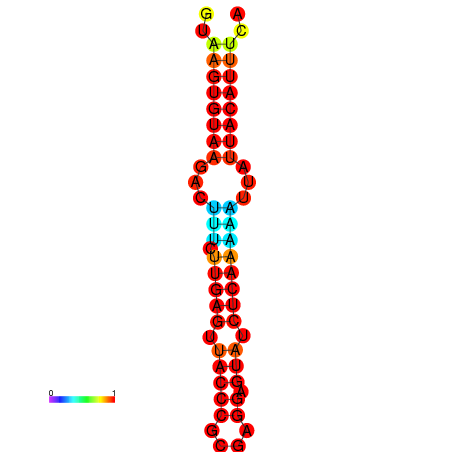 |
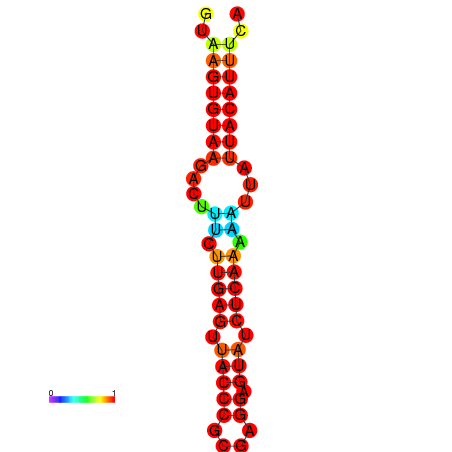 |
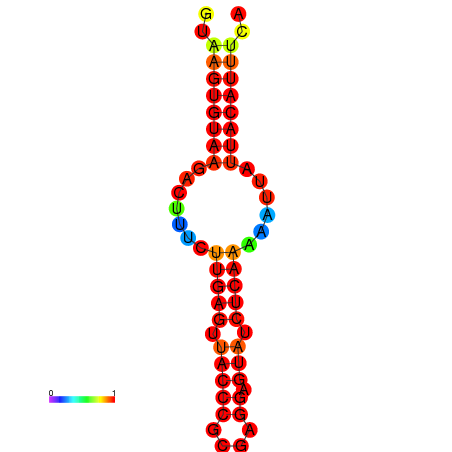 |
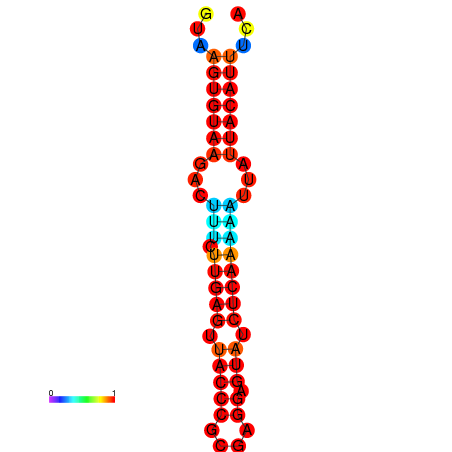 |
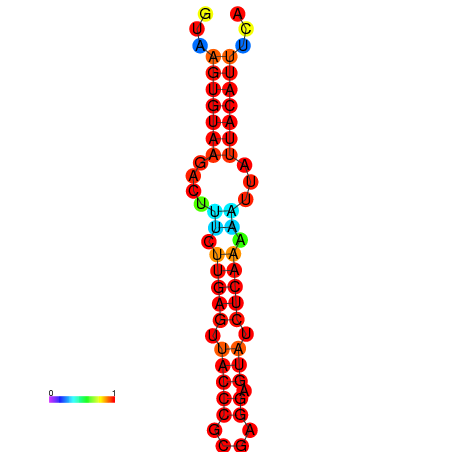 |
| dG=-11.5, p-value=0.009901 | dG=-10.8, p-value=0.009901 |
|---|---|
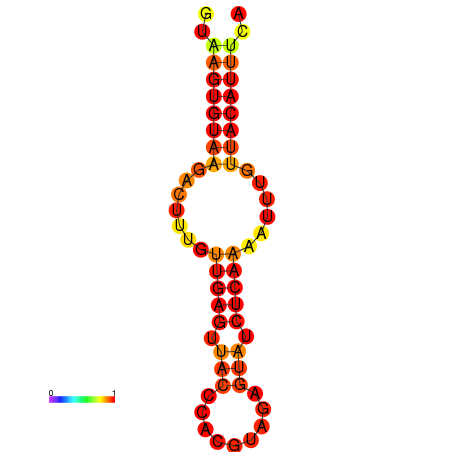 |
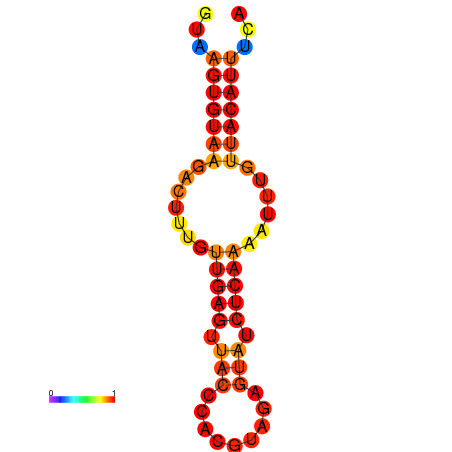 |
| dG=-3.8, p-value=0.009901 | dG=-3.4, p-value=0.009901 | dG=-3.2, p-value=0.009901 | dG=-2.8, p-value=0.009901 | dG=-2.8, p-value=0.009901 |
|---|---|---|---|---|
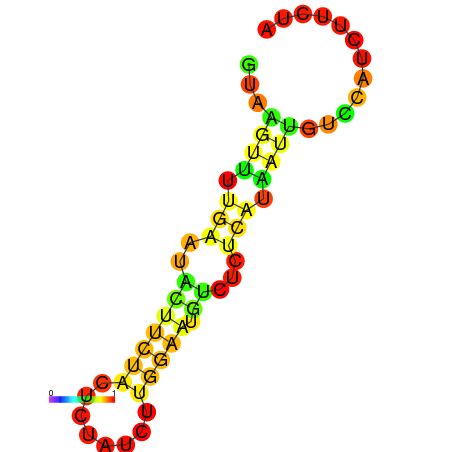 |
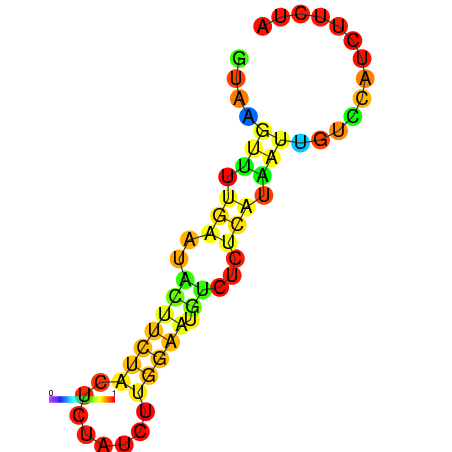 |
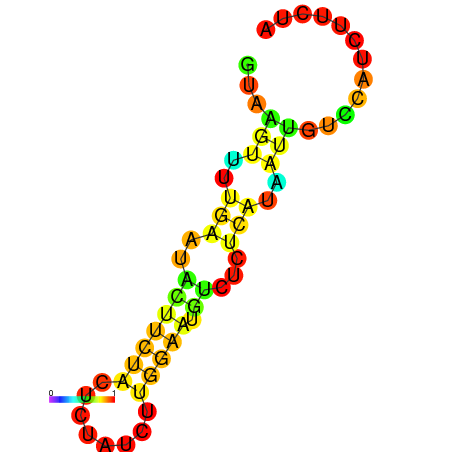 |
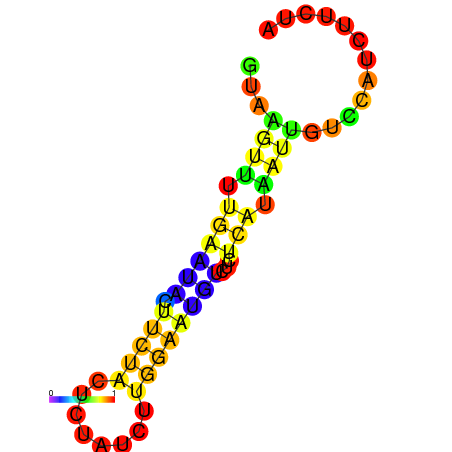 |
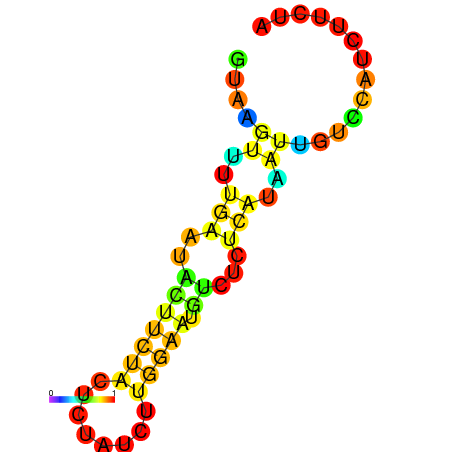 |
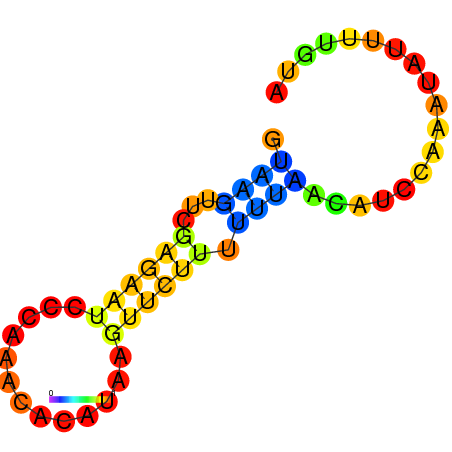
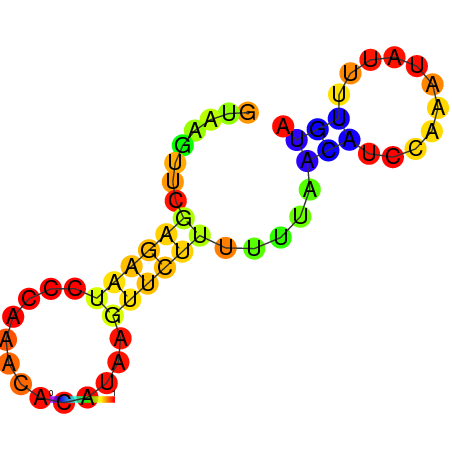
| dG=-1.7, p-value=0.009901 | dG=-1.2, p-value=0.009901 | dG=-0.9, p-value=0.009901 | dG=-0.7, p-value=0.009901 |
|---|---|---|---|
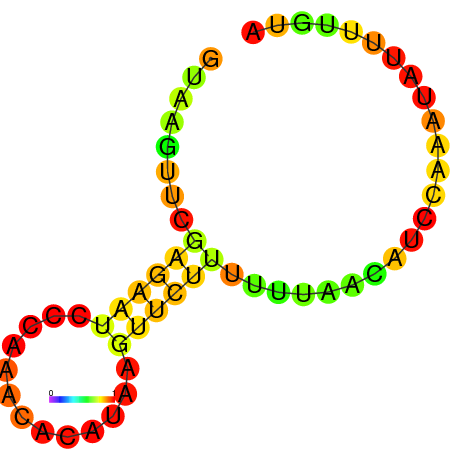 |
 |
 |
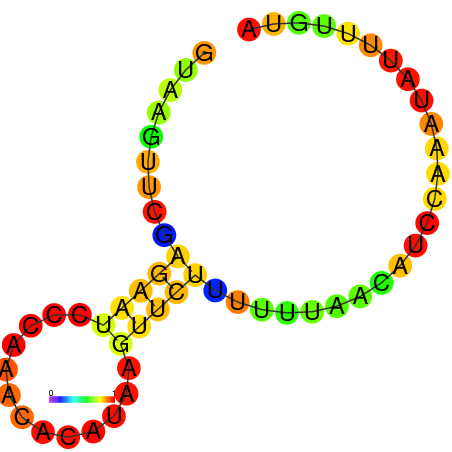 |
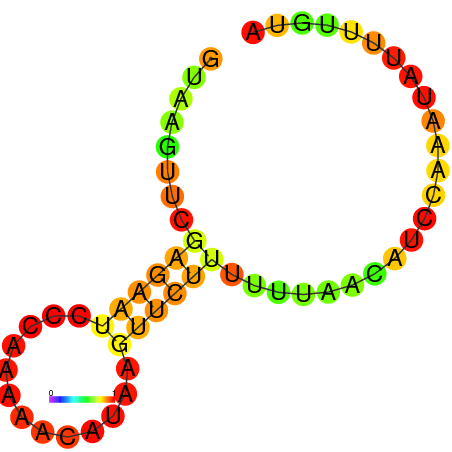
| dG=-1.7, p-value=0.009901 | dG=-1.2, p-value=0.009901 | dG=-0.9, p-value=0.009901 | dG=-0.7, p-value=0.009901 |
|---|---|---|---|
 |
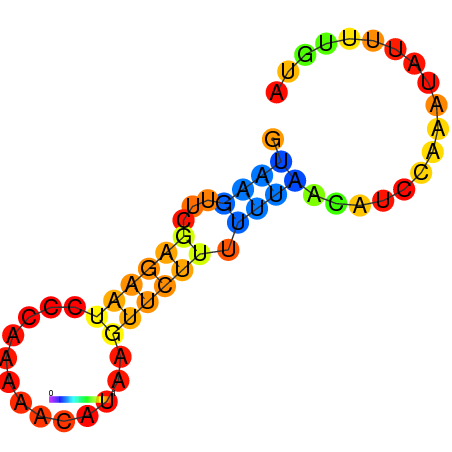 |
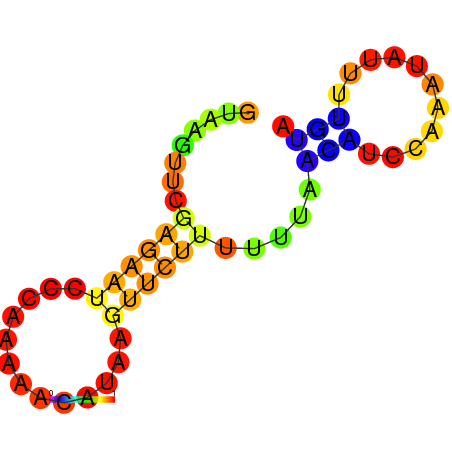 |
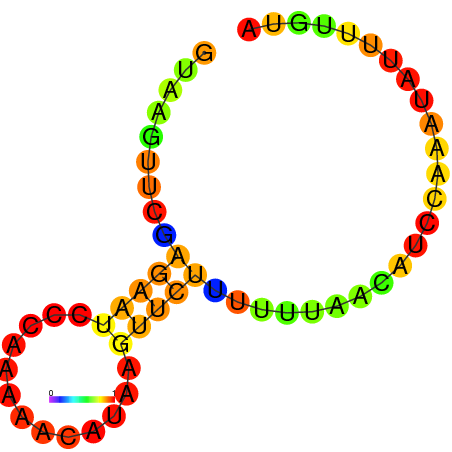 |
| dG=-7.7, p-value=0.009901 | dG=-7.3, p-value=0.009901 | dG=-6.7, p-value=0.009901 | dG=-6.2, p-value=0.009901 |
|---|---|---|---|
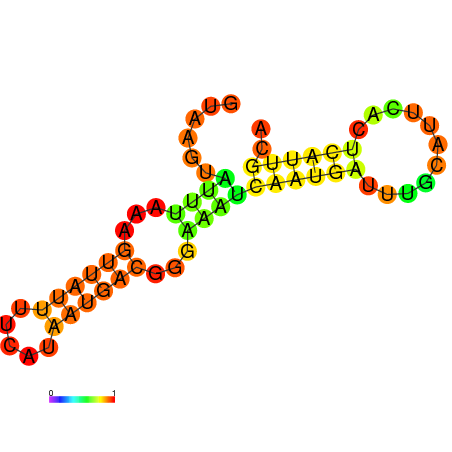 |
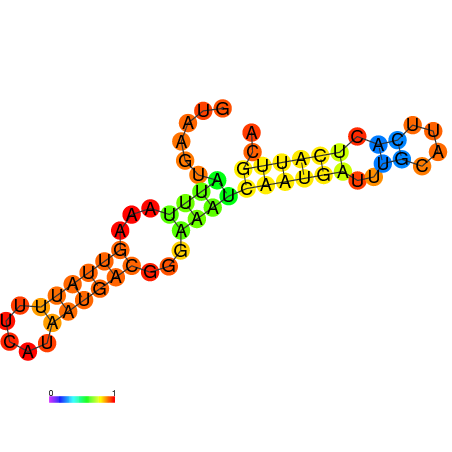 |
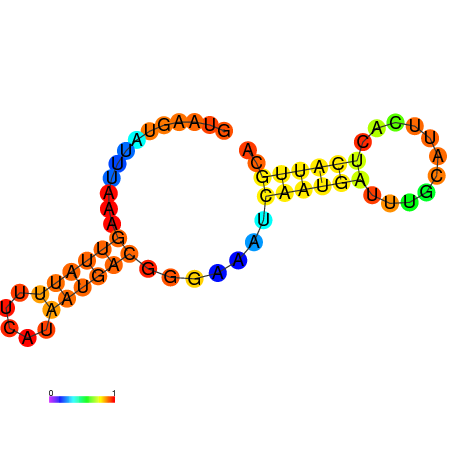 |
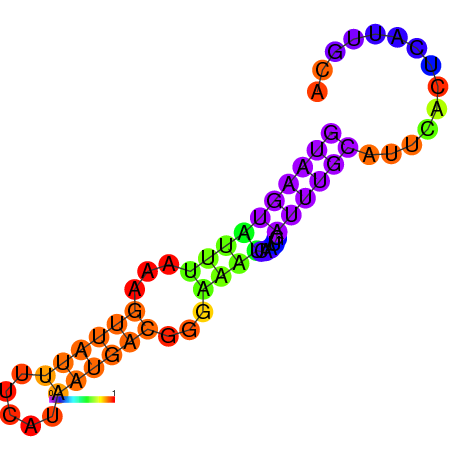 |
No secondary structure predictions returned
| dG=-4.6, p-value=0.009901 | dG=-4.5, p-value=0.009901 | dG=-3.8, p-value=0.009901 | dG=-3.8, p-value=0.009901 | dG=-3.6, p-value=0.009901 |
|---|---|---|---|---|
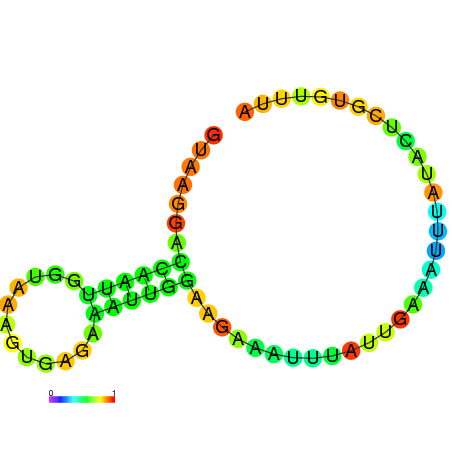 |
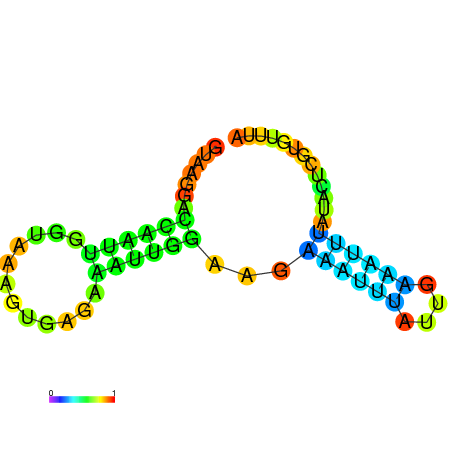 |
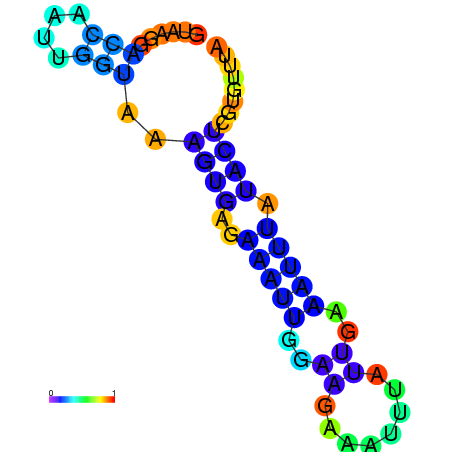 |
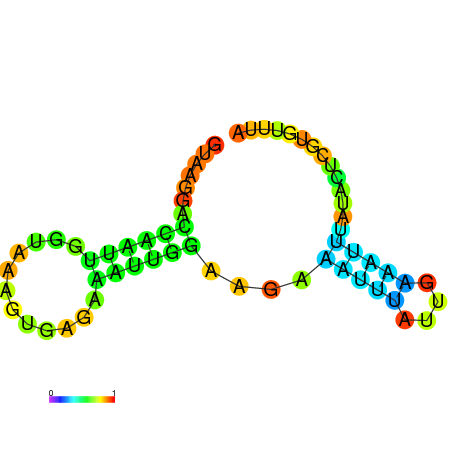 |
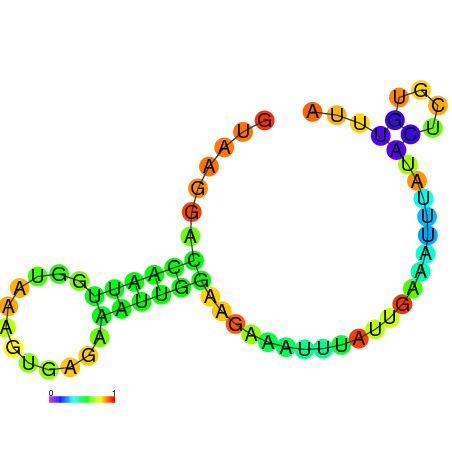 |
No secondary structure predictions returned
Generated: 03/07/2013 at 04:46 PM