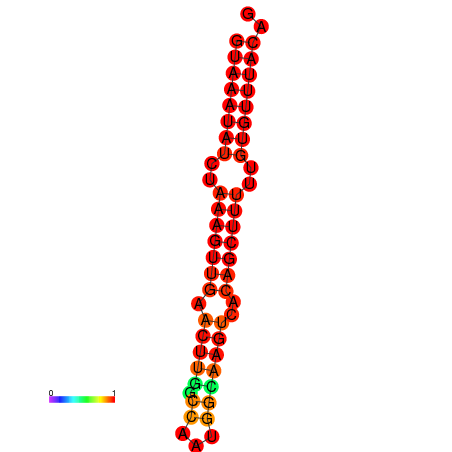
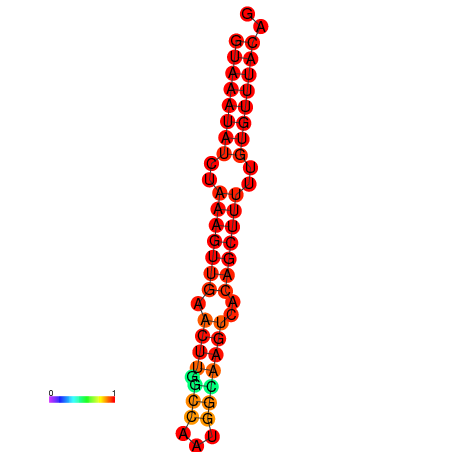
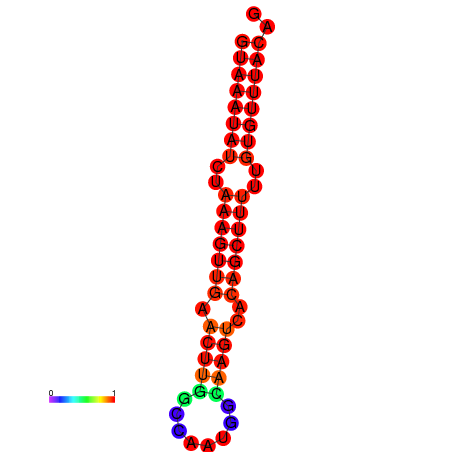
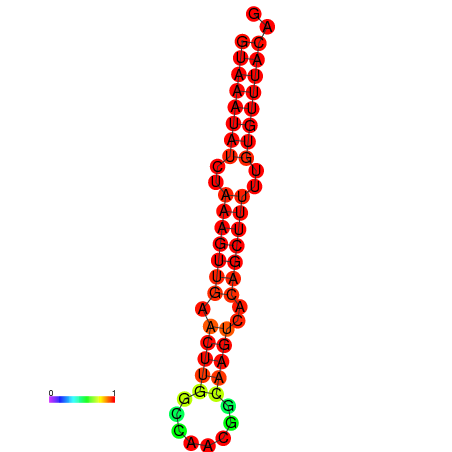
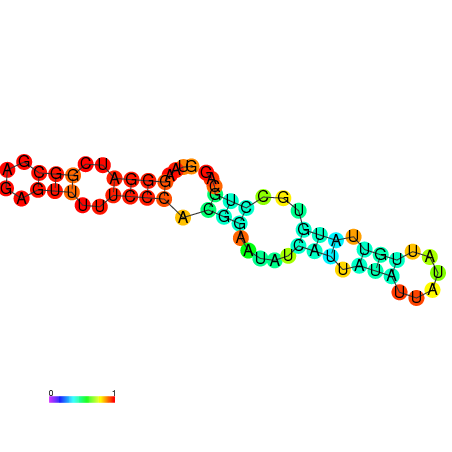
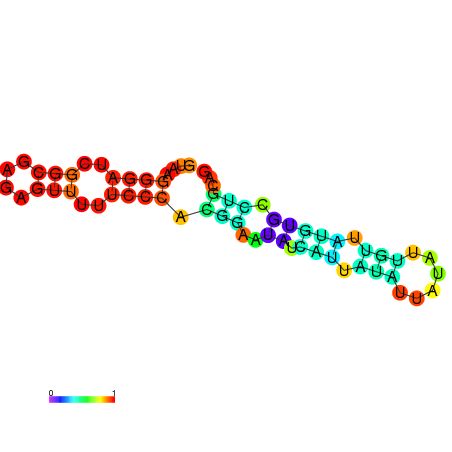
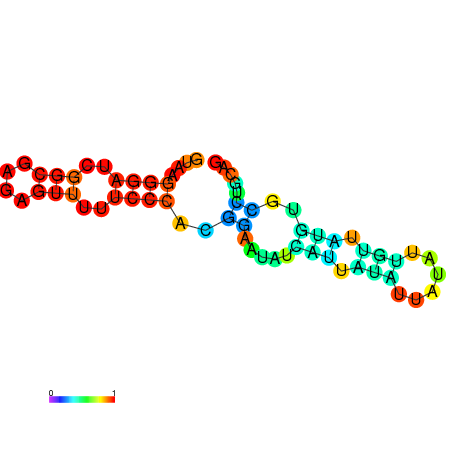
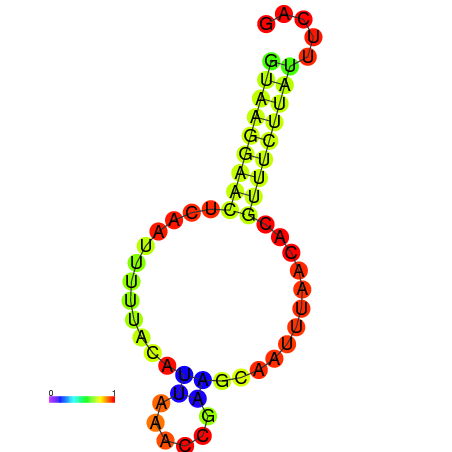| dm3 |
chr2R:6773023-6773109 + |
AAGTTCCTCATCTATGTAAATA-TCTAAA-------------------GTTGAACTTGGCCAATG-------------------------------GCAAGTCACAGCTTTTTGTGTTTACAGGAGAACGAGTACTTC |
| droSim1 |
chr2R:5362602-5362688 + |
AAGTTCCTCATCTATGTAAATA-TCTAAA-------------------GTTGAACTTGGCCAACG-------------------------------GCAAGTCACAGCTTTTTGTGTTTACAGGAGAACGAGTACTTC |
| droSec1 |
super_1:4360092-4360178 + |
AAGTTCCTCATCTATGTAAATA-TCTAAA-------------------GTTGAACTTGGCCAACG-------------------------------GCAAGTCACAGCTTTTTGTGTTTACAGGAGAACGAGTACTTC |
| droYak2 |
chr2R:2355901-2355987 - |
AAGTTCCTGATATACGTAAATA-TCTAAA-------------------GTTGAACTTGGCCAACG-------------------------------GCAAGTCACAGCTTTTTGTGTTTACAGGAGAACGAATACTTC |
| droEre2 |
scaffold_4845:19001852-19001938 - |
AAGTTCCTCATATACGTAAATA-TCTAAA-------------------GTTGAACTTGGCCAACG-------------------------------GCAAGTCACAGCTTTTTGTGTTTACAGGAGAACGAGTACTTC |
| droAna3 |
scaffold_13266:18269903-18269989 + |
AAGTTTCTTATCTACGTAAGGA-ACTCAA-------------------TTTT------------------------------TACATTAAACCGAAGCAATTTAACACGTTTCTTATT-TCAGGAGAACGAGTACTTC |
| dp4 |
chr3:12187375-12187468 + |
AAGTTTCTCATTTATGTAAGGG-ATCGGC-------------------G-AGAGTTTTTCCCACG-GA----ATA---TCATTATATT---------------ATATTGTTATGTGCCTGCAGGAAAACGAGTACTTT |
| droPer1 |
super_44:370822-370915 + |
AAGTTTCTCATTTATGTAAGGG-ATCGGC-------------------G-AGAGTTTTTCCCACG-GA----ATA---TCATTATATT---------------ATATTGTTATGTGCCTGCAGGAAAACGAGTACTTT |
| droWil1 |
scaffold_180697:3635906-3636001 - |
AAGTTTCTTATCTATGTAAGACAATCCAC-------------------G-AGGTGGTT------GGGAAGACAAATTTTGAATA-ATT---------------GGATTTTGATTTTTCTACAGGAAAACGAATATTTT |
| droVir3 |
scaffold_12875:11727599-11727677 + |
AAATTTCTCATCTATGTAAGTG-ATGA---------------------------TGCGTCCATTGGGA----ATA---TCATTT------------------------AATTTGTGTTGGTAGGAAAACGAATATTTT |
| droMoj3 |
scaffold_6496:15129877-15129967 - |
AAGTTTCTCATCTATGTAAGTA-GTAATA-------------------GGTGTTTGTAGACAT-----------AT--TCAG--------------TTAATTTTCGCATTTGTGTATTGGCAGGAAAACGAATATTTT |
| droGri2 |
scaffold_15245:6497403-6497490 - |
AAATTTCTCATCTATGTAAGTG-ATGCAAATGAATAAATATTAATCATATTGCCTTTAATCAATG-------------------------------GT------------------CTTGCAGGAGAATGAGTATTTC |