| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:21167397-21167491 - | ACGAATCTGTACAGTGTAAGCAGTGTTTGAACTCGATC--T-TGGTTC--TTGGA------C--TCTTGATAAGCTCAATTAACTGTTT----GCAGCGGACGGGATCTTCT |
| droSim1 | chrX:12920203-12920297 + | ACGAATCTGTACAGTGTAAGCAGTGTTTGAACTCGATC--T-TGGTTC--TTGGA------C--TCTTGATAAGCTCAATTAACTGTTT----GCAGCGGACGGCATCTTCT |
| droSec1 | super_8:3474404-3474498 - | ACGAATCTGTACAGTGTAAGCAGTGTTTGAACTCGATC--T-TGGTTC--TTGGA------C--TCTTGATAAGCTCAATTAACTGTTT----GCAGCGGACGGCATCTTCT |
| droYak2 | chrX:20343121-20343215 - | ACGAATCTGTTCAGTGTAAGCAGTGTTTGAACTCGATC--T-AGGATC--TTGGA------C--TCTTGATAAGCTCAATTAACTGTTT----GCAGCGGACGGCATCTTCT |
| droEre2 | scaffold_4690:17696754-17696848 - | ACGAATCTGTTCAGTGTAAGCAGTGTTTGAACTCGATC--T-GGGATC--TTGGA------C--TCTTGATAAGCTCAATTAACTGTTT----GCAGCGGACGGCATCTTCT |
| droAna3 | scaffold_13334:1054378-1054469 + | ACGAATCCGTACAGTGTAAGCAGTGTTTGAACTCGATC--T----------TGGAA--TAGC--TCCCGATAAGCTCAATTAACTGTTT----GCAGCCGATGGCATCTTCT |
| dp4 | chrXL_group1a:9060607-9060705 + | ACGAATCGATACAGTGTAAGCAGCGATTGA--TCAATCAAT-TGAATCGAATCGA------ATCGAATGATAAACTCCATTAACTGTTT----GCAGCCGATGGCATCTTCT |
| droPer1 | super_15:1454063-1454161 + | ACGAATCGATACAGTGTAAGCAGCGATTGA--TCAATCAAT-TGAATCGAATCGA------ATCGAATGATAAACTCCATTAACTGTTT----GCAGCCGATGGCATCTTCT |
| droWil1 | scaffold_180702:162895-162984 - | ATGAATCCATGCAATGTGAGTT--TTTTGATTTT---C--T-----TG--TTTAA------T--TTCGAAAAACTTTAATCGACTGTATTCTATCAGCTGATGGCATCTTCT |
| droVir3 | scaffold_12970:7925725-7925826 - | ACGAATCAATGAAATGTAAGCAGTGCTTGAGCTTATTC--TCTGGCTTCATTTGAC--CA--TTTTCTGATAAGCTCAATTAACTGTTT----GCAGCGGATGCCGCCCTCT |
| droMoj3 | scaffold_6482:1600635-1600734 + | ATGAGTCCATGAAATGTAAGCAGTGTTTGAACTAAATC--TCTGGCTA--CTTGGC--CG--TATATTGATAAGCTCAACTAACTGTTT----GCAGCGGACACCTATTTGG |
| droGri2 | scaffold_15203:11134564-11134665 + | GCGAATCAATACAATGTAAGCAGTGTTTGAACTGAATA--TCTGGTTA--TCCGACTTCA--TATACTGATAAGCTCAATTCACTGTTT----GCAGCGGATGGCGTCTTCT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:21167397-21167491 - | ACGAATCTGTACAGTGTAAGCAGTGTTTGAACTCGATC--T-TGGTTC--TTGGA----CT------CTTGATAAGCTCAATTAACTGTTTGCAGCGGACGGGATCTTCT |
| droSim1 | chrX:12920203-12920297 + | ACGAATCTGTACAGTGTAAGCAGTGTTTGAACTCGATC--T-TGGTTC--TTGGA----CT------CTTGATAAGCTCAATTAACTGTTTGCAGCGGACGGCATCTTCT |
| droSec1 | super_8:3474404-3474498 - | ACGAATCTGTACAGTGTAAGCAGTGTTTGAACTCGATC--T-TGGTTC--TTGGA----CT------CTTGATAAGCTCAATTAACTGTTTGCAGCGGACGGCATCTTCT |
| droYak2 | chrX:20343121-20343215 - | ACGAATCTGTTCAGTGTAAGCAGTGTTTGAACTCGATC--T-AGGATC--TTGGA----CT------CTTGATAAGCTCAATTAACTGTTTGCAGCGGACGGCATCTTCT |
| droEre2 | scaffold_4690:17696754-17696848 - | ACGAATCTGTTCAGTGTAAGCAGTGTTTGAACTCGATC--T-GGGATC--TTGGA----CT------CTTGATAAGCTCAATTAACTGTTTGCAGCGGACGGCATCTTCT |
| droAna3 | scaffold_13334:1054378-1054469 + | ACGAATCCGTACAGTGTAAGCAGTGTTTGAACTCGATC--T----------TGGAATAGCT------CCCGATAAGCTCAATTAACTGTTTGCAGCCGATGGCATCTTCT |
| dp4 | chrXL_group1a:9060607-9060705 + | ACGAATCGATACAGTGTAAGCAGCGATTGA--TCAATCAAT-TGAATCGAATCGA----AT----CGAATGATAAACTCCATTAACTGTTTGCAGCCGATGGCATCTTCT |
| droPer1 | super_15:1454063-1454161 + | ACGAATCGATACAGTGTAAGCAGCGATTGA--TCAATCAAT-TGAATCGAATCGA----AT----CGAATGATAAACTCCATTAACTGTTTGCAGCCGATGGCATCTTCT |
| droVir3 | scaffold_12970:7925725-7925826 - | ACGAATCAATGAAATGTAAGCAGTGCTTGAGCTTATTC--TCTGGCTTCATTTGA----CC--ATTTTCTGATAAGCTCAATTAACTGTTTGCAGCGGATGCCGCCCTCT |
| droMoj3 | scaffold_6482:1600635-1600734 + | ATGAGTCCATGAAATGTAAGCAGTGTTTGAACTAAATC--TCTGGCTA--CTTGG----CC--GTATATTGATAAGCTCAACTAACTGTTTGCAGCGGACACCTATTTGG |
| droGri2 | scaffold_15203:11134564-11134665 + | GCGAATCAATACAATGTAAGCAGTGTTTGAACTGAATA--TCTGGTTA--TCCGA----CTTCATATACTGATAAGCTCAATTCACTGTTTGCAGCGGATGGCGTCTTCT |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
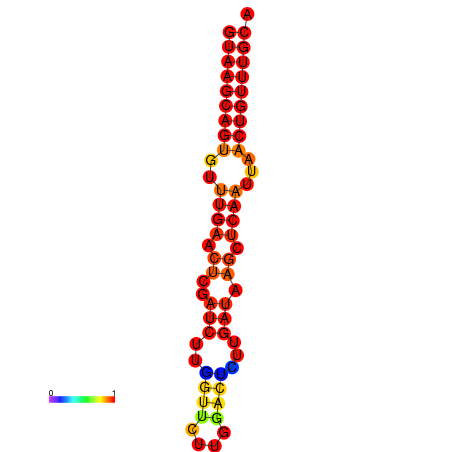
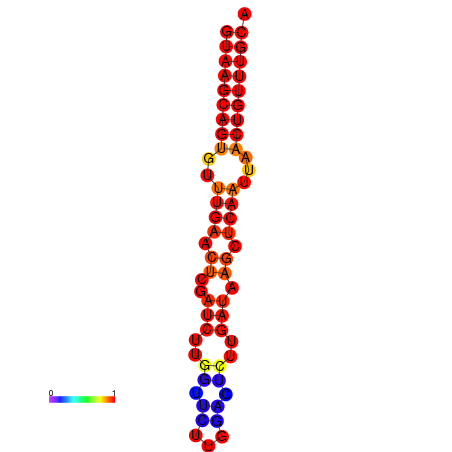
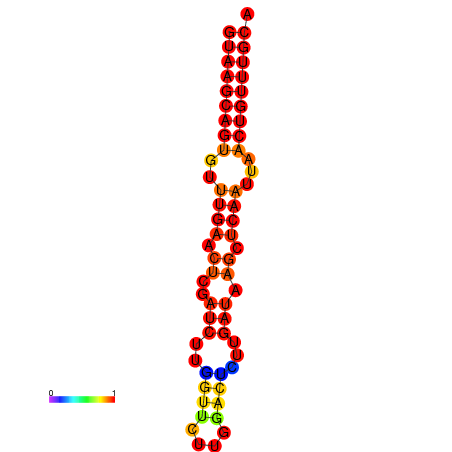
| dG=-16.8, p-value=0.009901 | dG=-16.6, p-value=0.009901 |
|---|---|
 |
 |
| dG=-16.8, p-value=0.009901 | dG=-16.6, p-value=0.009901 |
|---|---|
 |
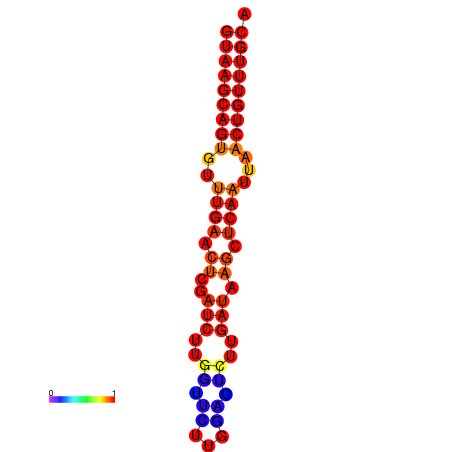 |
| dG=-16.8, p-value=0.009901 | dG=-16.6, p-value=0.009901 |
|---|---|
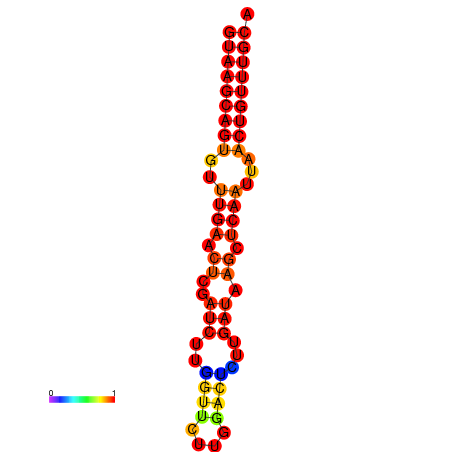 |
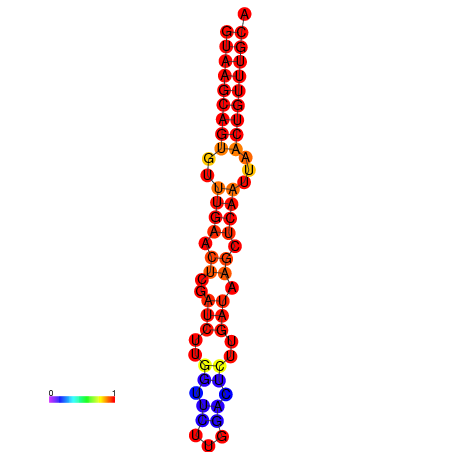 |
| dG=-17.8, p-value=0.009901 | dG=-17.3, p-value=0.009901 | dG=-17.1, p-value=0.009901 |
|---|---|---|
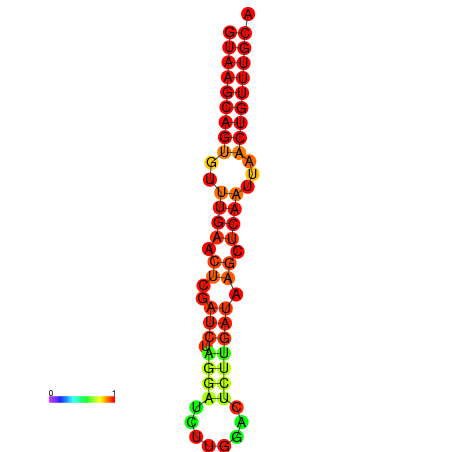 |
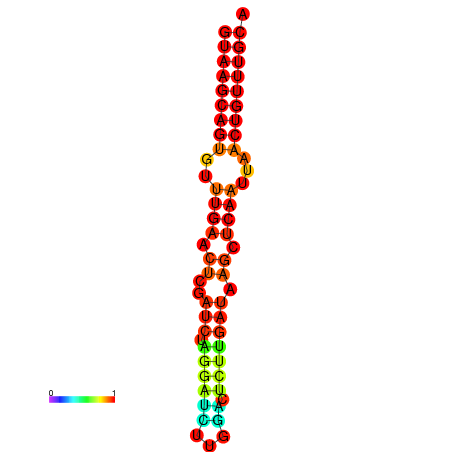 |
 |
| dG=-17.3, p-value=0.009901 | dG=-17.2, p-value=0.009901 | dG=-17.1, p-value=0.009901 | dG=-17.0, p-value=0.009901 | dG=-16.7, p-value=0.009901 |
|---|---|---|---|---|
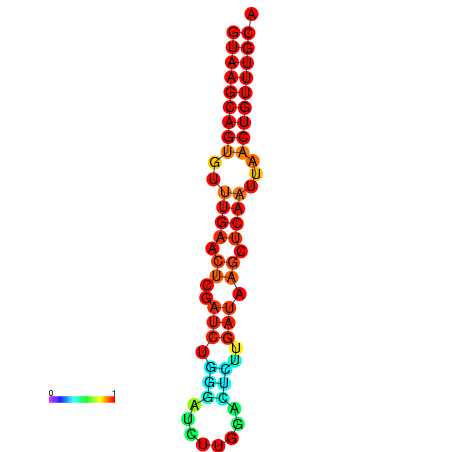 |
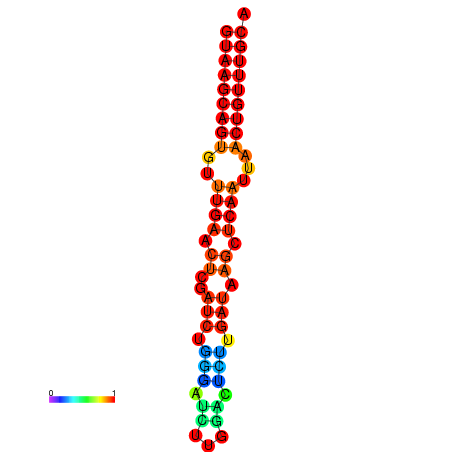 |
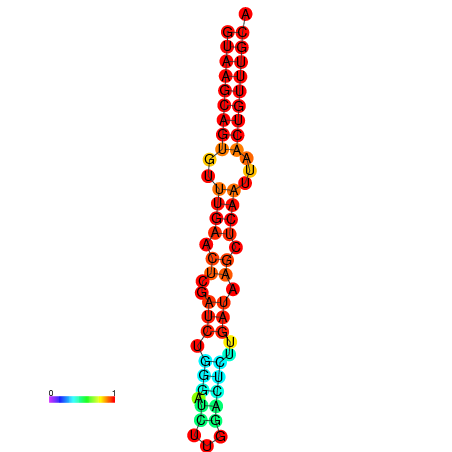 |
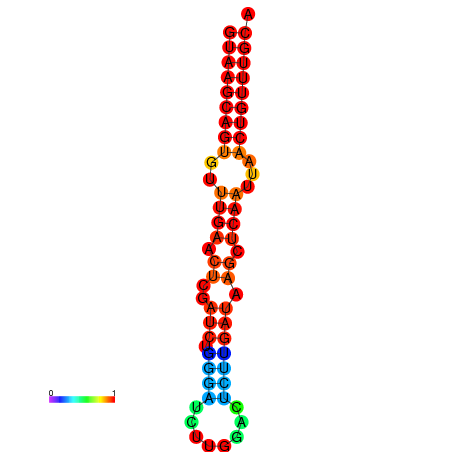 |
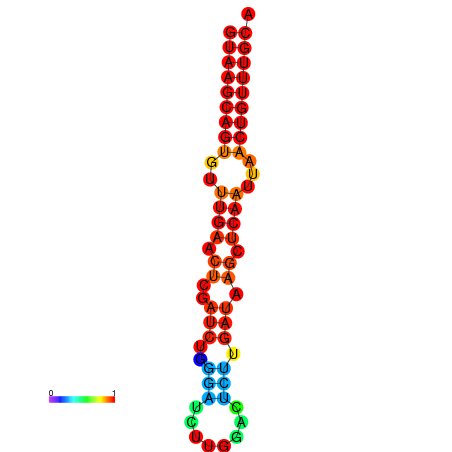 |
| dG=-17.8, p-value=0.009901 |
|---|
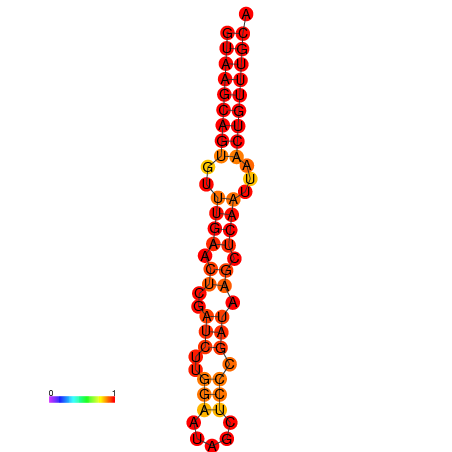 |
| dG=-16.0, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.4, p-value=0.009901 | dG=-15.2, p-value=0.009901 |
|---|---|---|---|
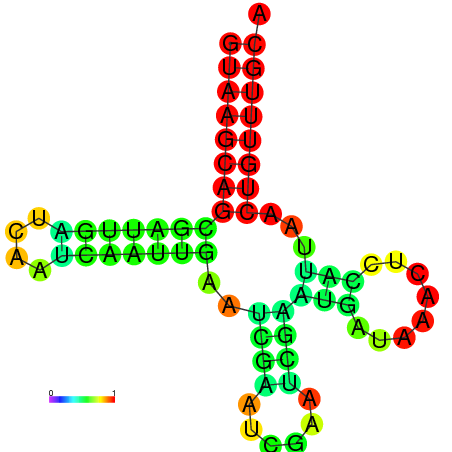 |
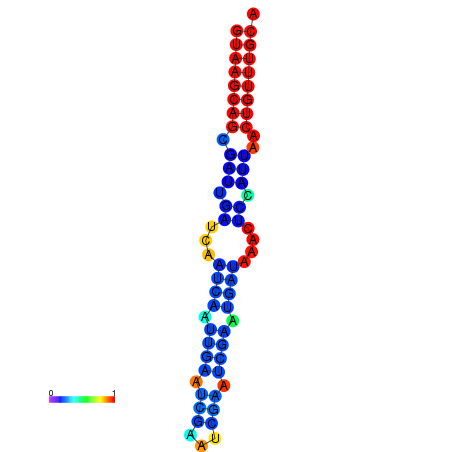 |
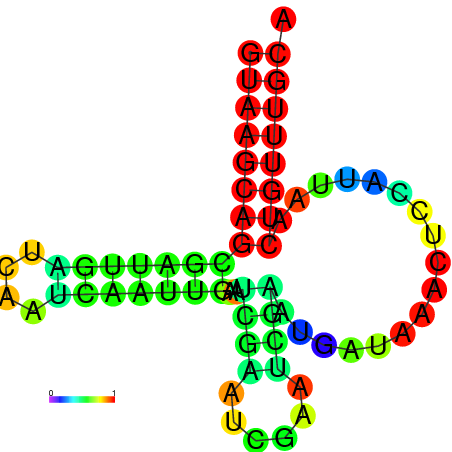 |
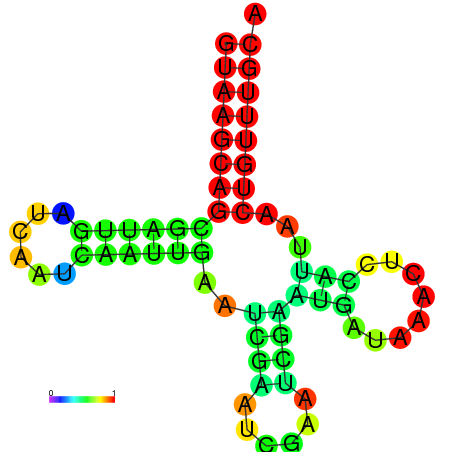 |
| dG=-16.0, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.4, p-value=0.009901 | dG=-15.2, p-value=0.009901 |
|---|---|---|---|
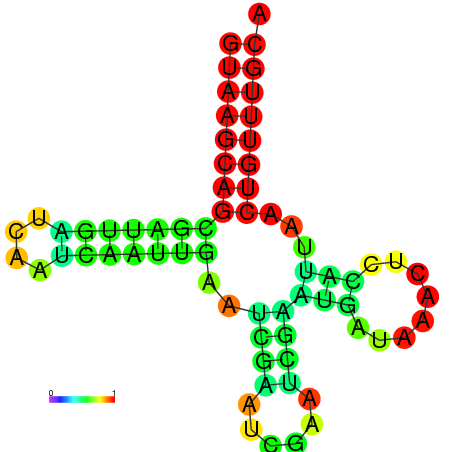 |
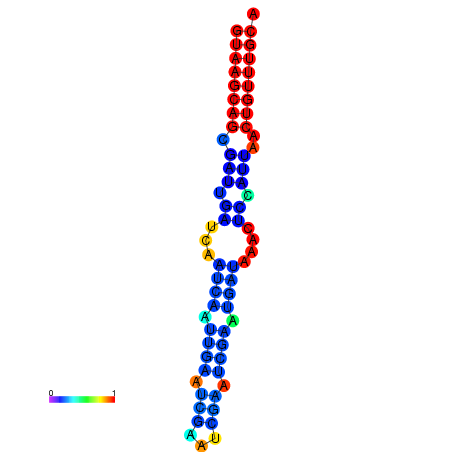 |
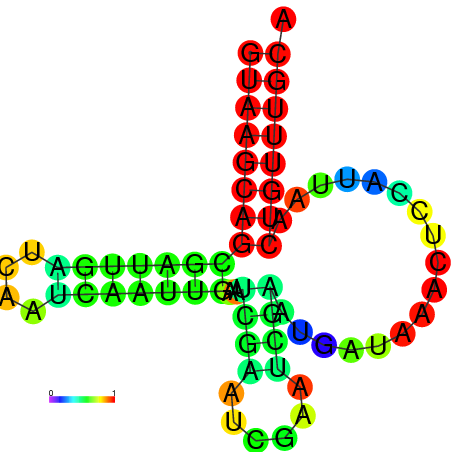 |
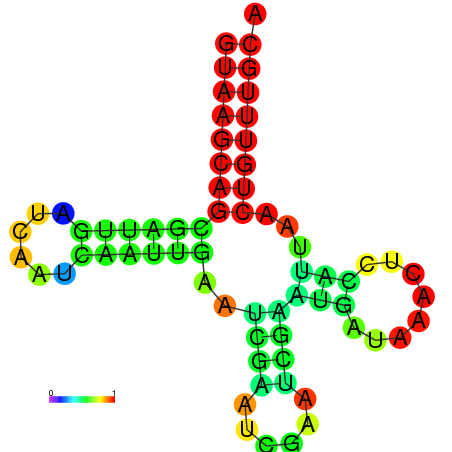 |
| dG=-8.7, p-value=0.009901 | dG=-8.7, p-value=0.009901 | dG=-7.8, p-value=0.009901 | dG=-7.8, p-value=0.009901 | dG=-7.7, p-value=0.009901 |
|---|---|---|---|---|
 |
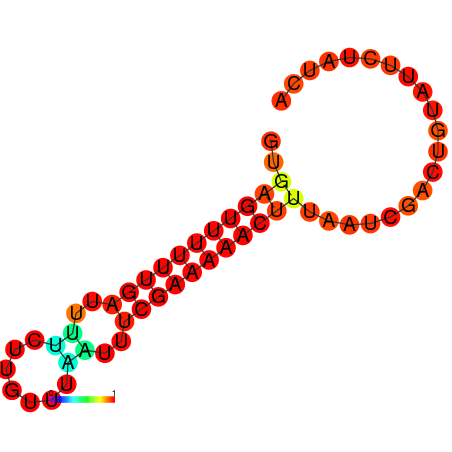 |
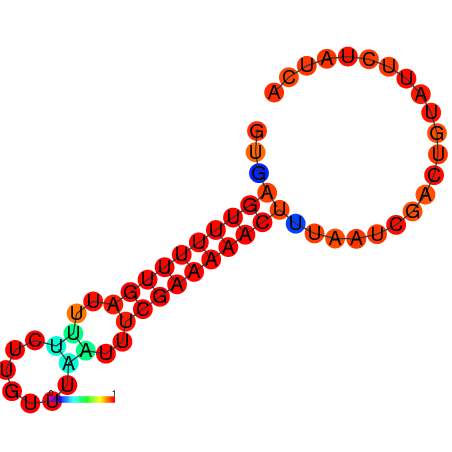 |
 |
 |
| dG=-24.5, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.5, p-value=0.009901 |
|---|---|---|
 |
 |
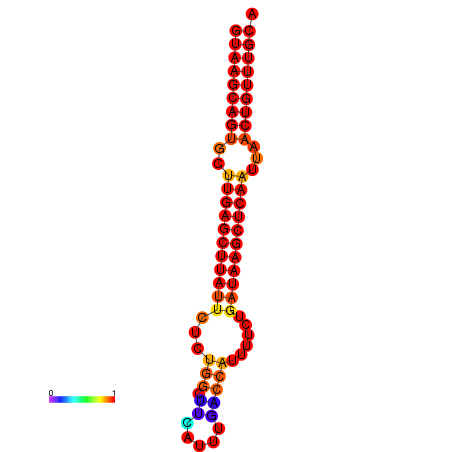 |
| dG=-18.3, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|
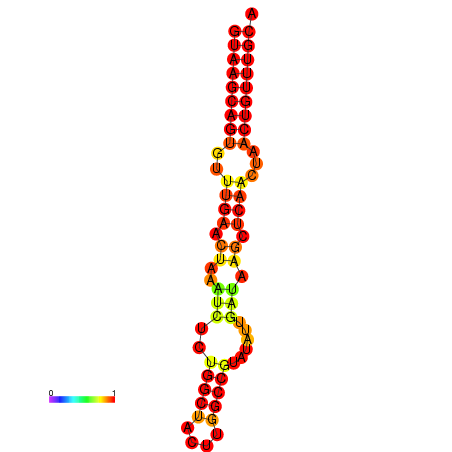 |
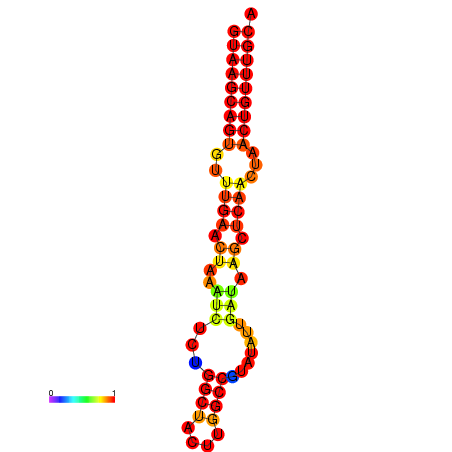 |
| dG=-17.8, p-value=0.009901 | dG=-17.7, p-value=0.009901 | dG=-17.0, p-value=0.009901 | dG=-16.9, p-value=0.009901 |
|---|---|---|---|
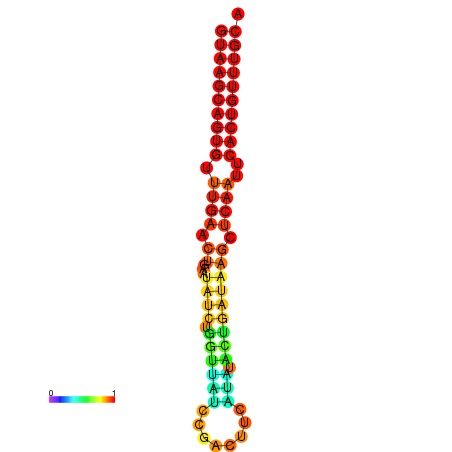 |
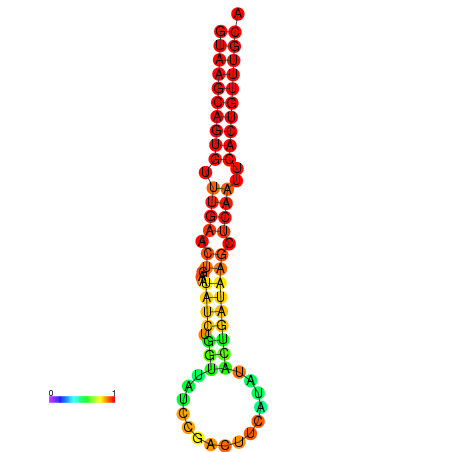 |
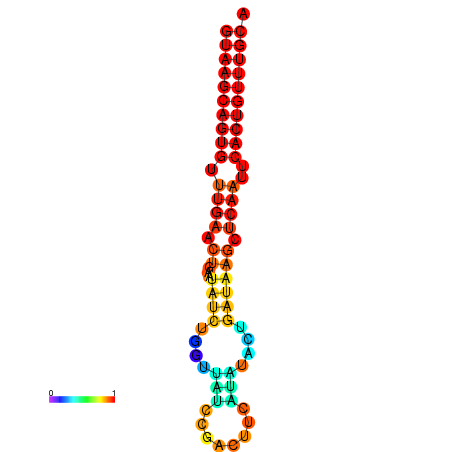 |
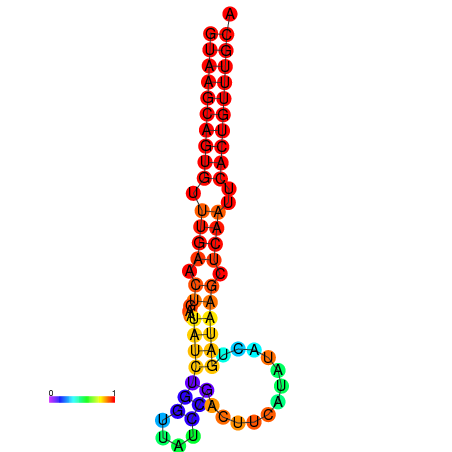 |
Generated: 03/07/2013 at 04:46 PM