| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:16724780-16724873 - | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droSim1 | chr2L:16422879-16422972 - | CGGGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droSec1 | super_7:371570-371663 - | CGGGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droYak2 | chr2R:4378132-4378225 + | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droEre2 | scaffold_4845:19796345-19796438 - | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droAna3 | scaffold_12943:3938325-3938418 + | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTCCAATACCAGCTG |
| dp4 | chr4_group3:3057660-3057753 + | CGCGTCAACTTTCAGGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--TGCCAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGCTG |
| droPer1 | super_1:4544383-4544476 + | CGCGTCAACTTTCAGGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--TGCCAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGCTG |
| droWil1 | scaffold_180708:1783971-1784066 + | CGTGTTAATTTCCAGGTGAGTTTGAAATTGAAATGCCTAAATTGTTTTGTGTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATATCAATTA |
| droVir3 | scaffold_12963:6536524-6536617 - | CGTGTCAACTTTCAGGTGAGTTTGAAATTGAAATATGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droMoj3 | scaffold_6500:25691037-25691130 - | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droGri2 | scaffold_15252:9753534-9753627 - | CGCGTCAATTTCCAAGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--GTCCAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGCTG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:16724780-16724873 - | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droSim1 | chr2L:16422879-16422972 - | CGGGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droSec1 | super_7:371570-371663 - | CGGGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droYak2 | chr2R:4378132-4378225 + | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droEre2 | scaffold_4845:19796345-19796438 - | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droAna3 | scaffold_12943:3938325-3938418 + | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGCGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTCCAATACCAGCTG |
| dp4 | chr4_group3:3057660-3057753 + | CGCGTCAACTTTCAGGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--TGCCAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGCTG |
| droPer1 | super_1:4544383-4544476 + | CGCGTCAACTTTCAGGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--TGCCAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGCTG |
| droWil1 | scaffold_180708:1783971-1784066 + | CGTGTTAATTTCCAGGTGAGTTTGAAATTGAAATGCCTAAATTGTTTTGTGTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATATCAATTA |
| droVir3 | scaffold_12963:6536524-6536617 - | CGTGTCAACTTTCAGGTGAGTTTGAAATTGAAATATGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droMoj3 | scaffold_6500:25691037-25691130 - | CGTGTCAACTTCCAGGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--GTACAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGTTG |
| droGri2 | scaffold_15252:9753534-9753627 - | CGCGTCAATTTCCAAGTGAGTTTGAAATTGAAATGTGTAAATTGTTTG--GTCCAATTTAAATTCGATTTCTTATTCATAGGTGCAATACCAGCTG |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
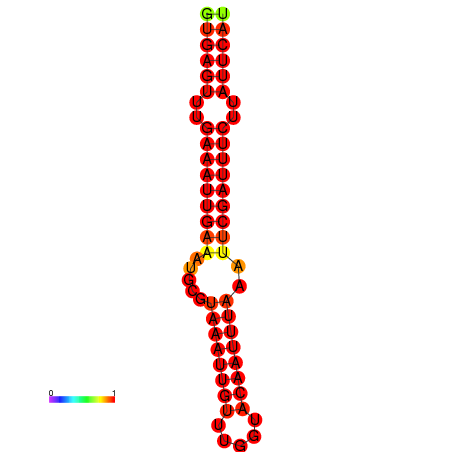
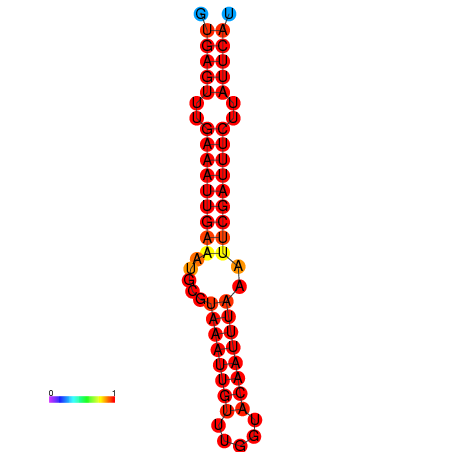
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
 |
 |
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
 |
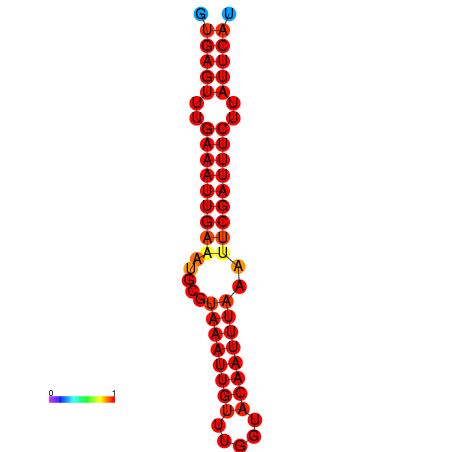 |
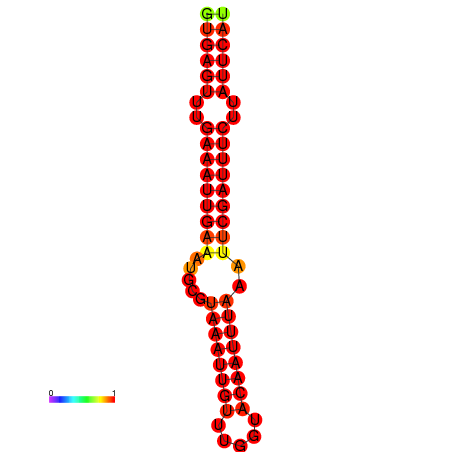
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
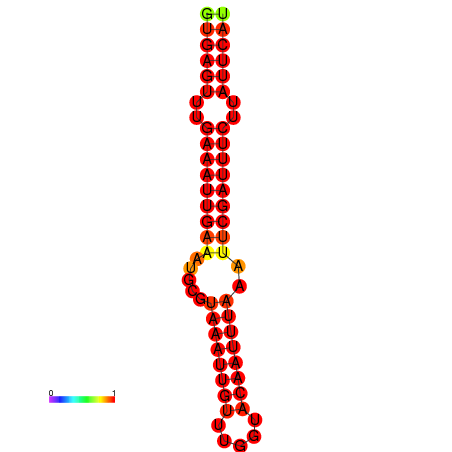 |
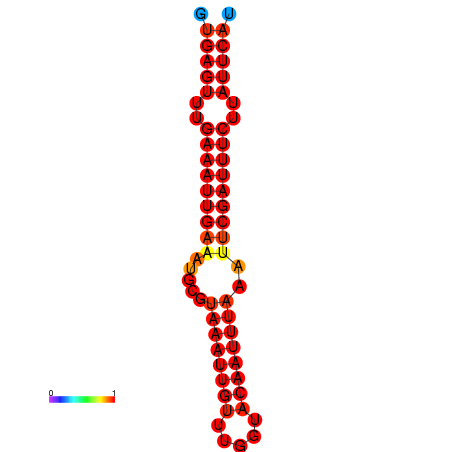 |
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
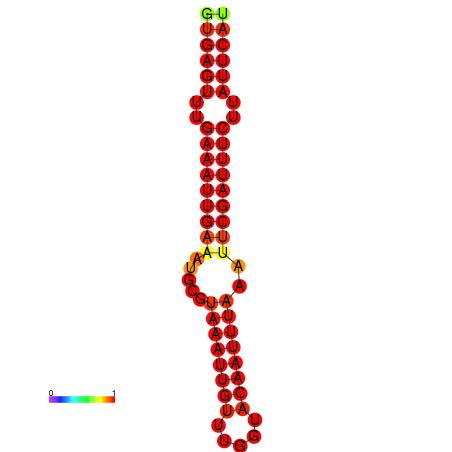 |
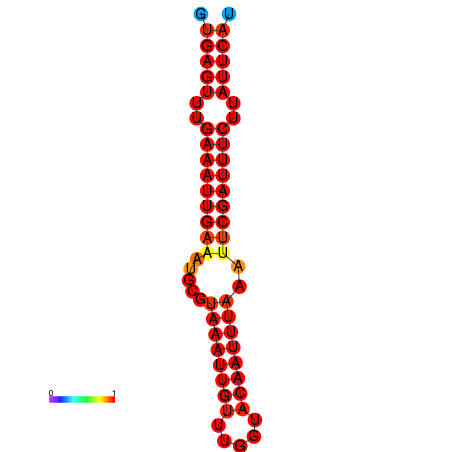 |
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
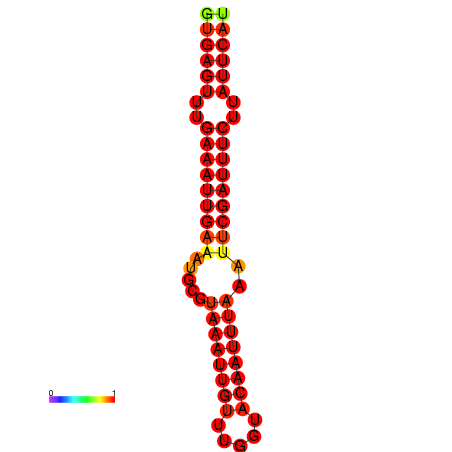 |
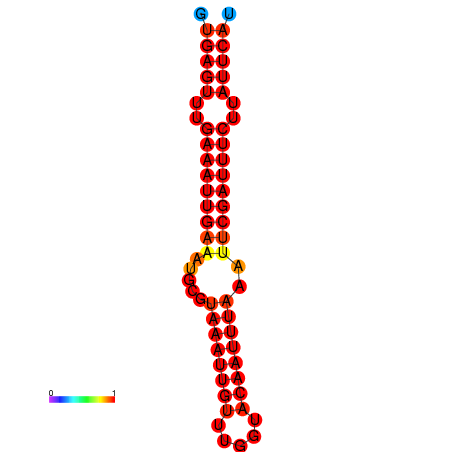 |
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
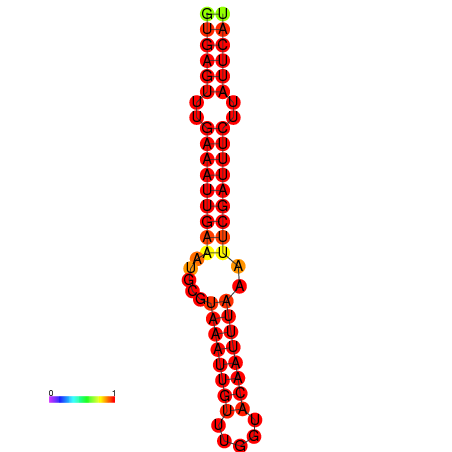 |
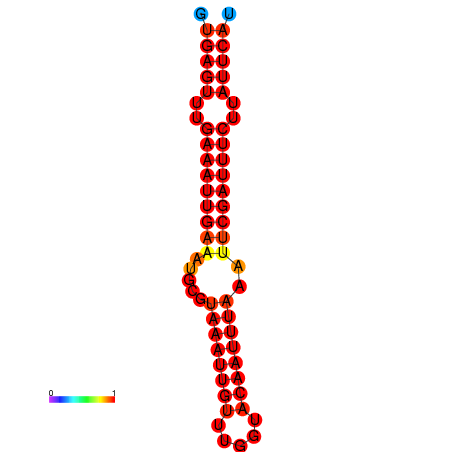 |
| dG=-16.1, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.1, p-value=0.009901 |
|---|---|---|
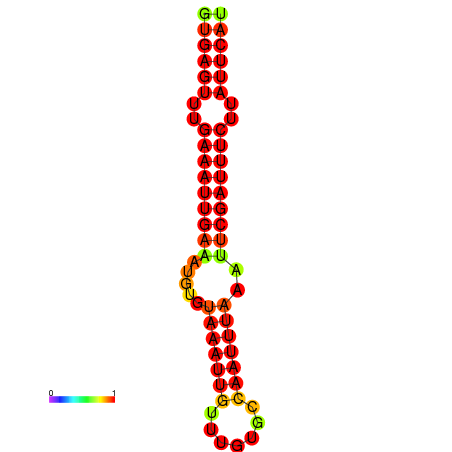 |
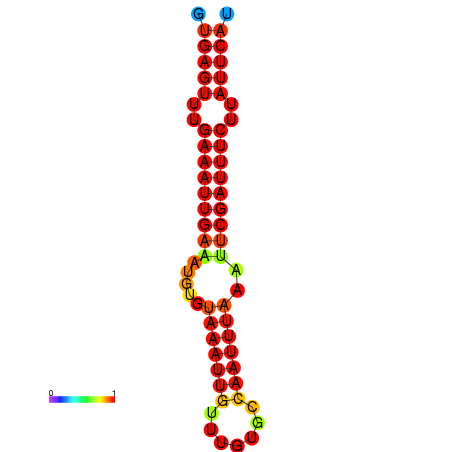 |
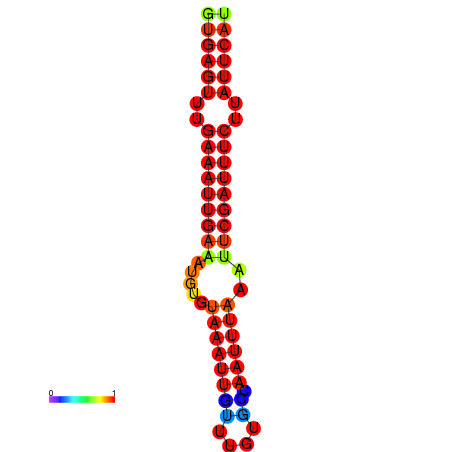 |
| dG=-16.1, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.1, p-value=0.009901 |
|---|---|---|
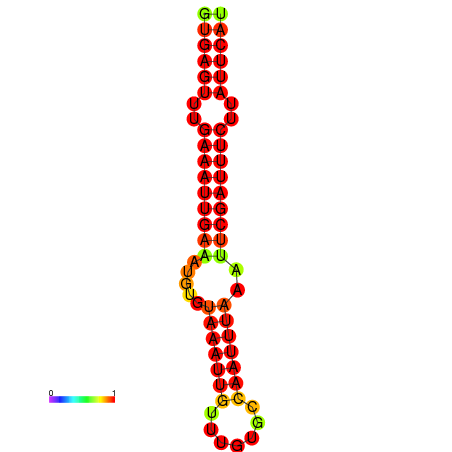 |
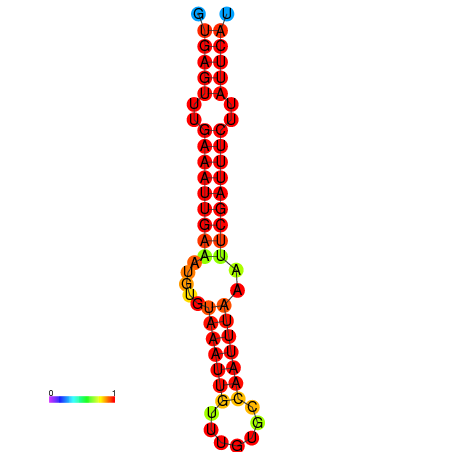 |
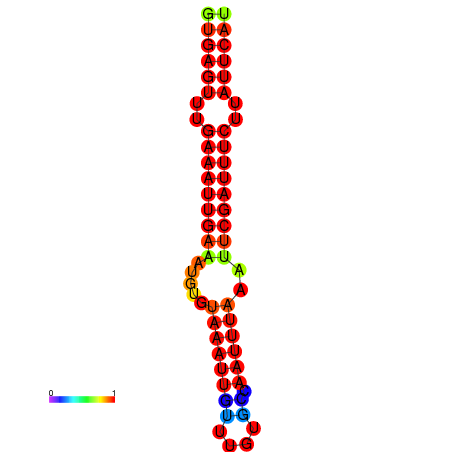 |
| dG=-17.4, p-value=0.009901 | dG=-16.8, p-value=0.009901 |
|---|---|
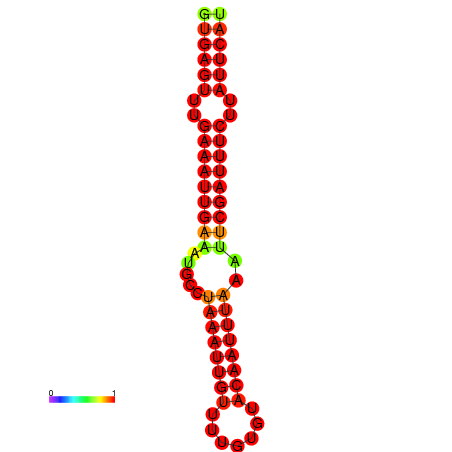 |
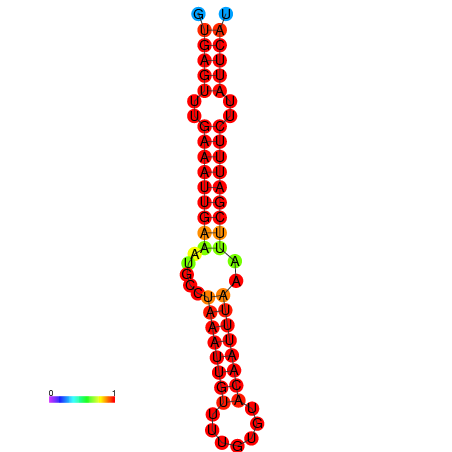 |
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
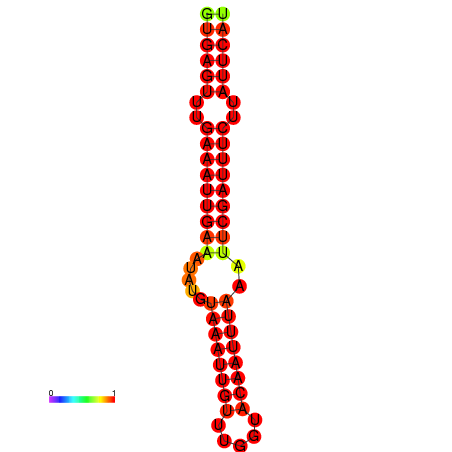 |
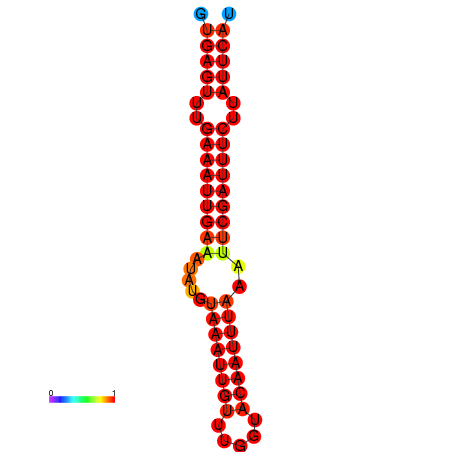 |
| dG=-18.5, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|
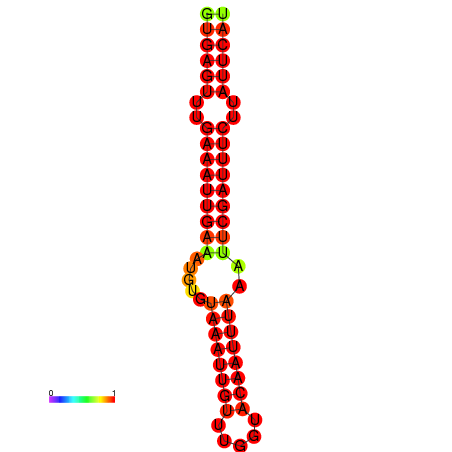 |
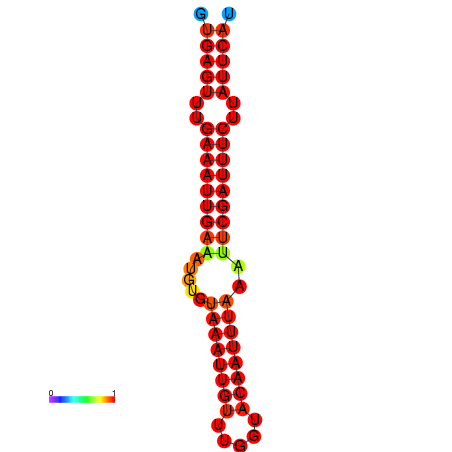 |
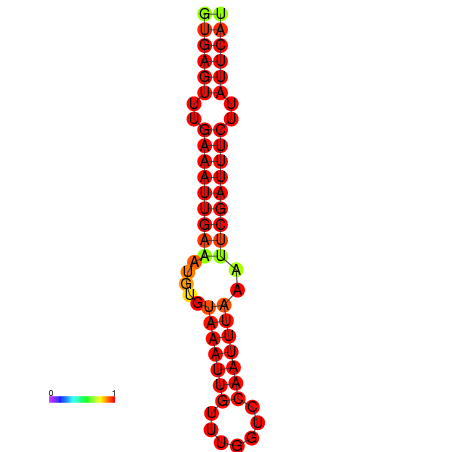
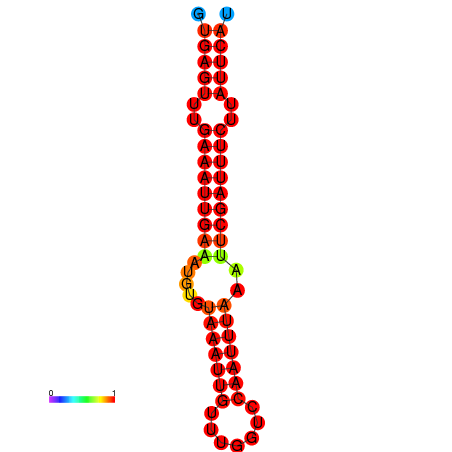
| dG=-16.1, p-value=0.009901 | dG=-15.5, p-value=0.009901 |
|---|---|
 |
 |
Generated: 03/07/2013 at 04:45 PM