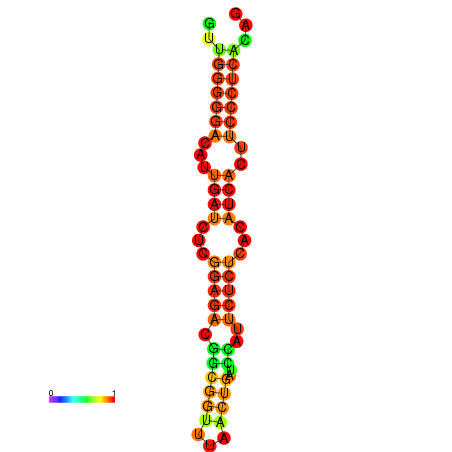
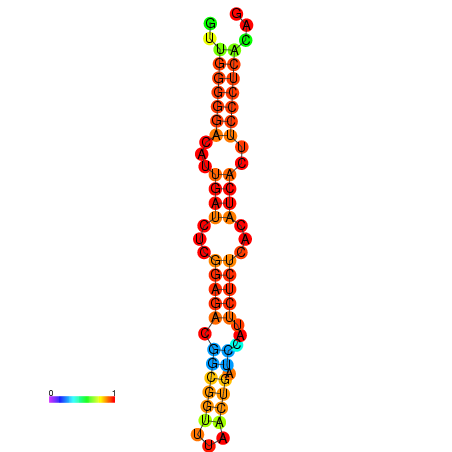
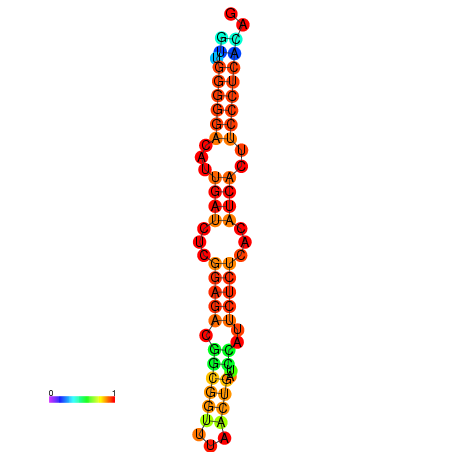
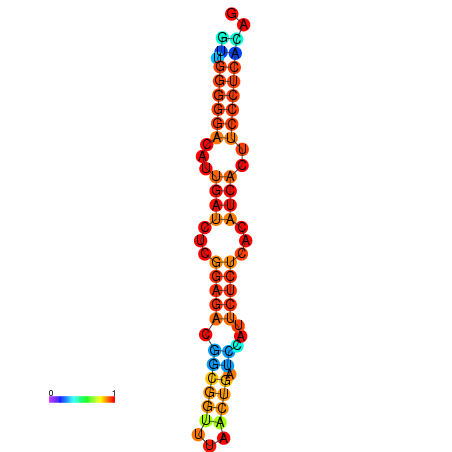
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:3767606-3767703 + | GTGGCTGGACAAAAGGT-TGGGGG--ACATTGATCTCGGAGACG-------------G-------------------------CGGTTTAACT--------------------GATCCATTCTCTCACATCACTT---CCCTCACAGTGCACGGTGCTTGTA |
| droSim1 | chr2L:3722744-3722841 + | GTGGCTGGACAAGAGGT-TGGGGG--ACATTGATCTCGGAGACG-------------G-------------------------CGGTTTAACT--------------------GATCCATTCTCTCACATCACTT---CCCTCACAGTGCGCGGTGCTTGTA |
| droSec1 | super_5:1870685-1870782 + | GTGGCTGGACAAGAGGT-TGGGGG--ACATTGATCTCTGAGACG-------------G-------------------------CGGTTTAACT--------------------GATCCATTCTCTCACATCACTT---CCCTCACAGTGCGCGGTGCTTGTA |
| droYak2 | chr2L:3769291-3769388 + | GTGGCTGGACAAGAGGT-TGGGGG--ACATTGATCTCGGAGACG-------------G-------------------------CGGTTTAACT--------------------GATCCATTCTCTCACATCACTT---CCCTCACAGTGCGCGATGTTTGTA |
| droEre2 | scaffold_4929:3814030-3814127 + | GTGGCTGGACAAGAGGT-TGGGGG--ACATTGATCTCGGAGACG-------------G-------------------------CGGTTTAACT--------------------GATCCAATCTCTCACATCACTT---CCCTCACAGTGCGCGGTGCTTGTA |
| droAna3 | scaffold_12916:5837615-5837739 + | GTGGCTGGACAAGAGGT-GAGATAACACGTT-----CTAACCAACTGATATGAAACTA------CGAATGTTTTTATCGCTGTCGATTCGAAC--------------------GA-----TCTCTCATATAACCGTACCTATTACAGTGCGCGGTGTCTGTA |
| dp4 | chr4_group3:4962038-4962146 - | GTGGCTGGACAAGAGGTGTGTGGG--ATACTGATTTTAGAGAAAAAAAAA----------A----------------------CCAT-T--AACGTGAGG--------------CTTCG-TTTCTCACATCATTT-TCCCCTCACAGTGCCCGGTGCCTGTA |
| droPer1 | super_1:6454682-6454788 - | GTGGCTGGACAAGAGGTGTGTGGG--ATACTGATTTTAGAGAAAA------------ATAA----------------------CCAT-T--AACGTGAGG--------------CTTCG-TTTCTCACATCATTT-TCCCCTCACAGTGCCCGGTGCCTGTA |
| droWil1 | scaffold_180708:9298763-9298869 - | ATGGTTGGATAAAAGGT--GGGGT--ACAACAACAACAAAAAT--------------A-------------------------CGAATATAATGGT-AAACTCTC-------TCTCTCAT---CTCTTATCATTT-TCTCCTCACAGTGCGCGTTCACTTTA |
| droVir3 | scaffold_12963:14941260-14941364 - | GTGGCTGGACAAAAGGT-TGGGGC--ACATTGATCTTCAAGAAACTCACAGCCA---G-------------------------CTCATT------------------------TACTCACTCTCTCTC-TCTTCT-CTCCCTCACAGTGCCCGCTGTCTGTA |
| droMoj3 | scaffold_6500:17898451-17898546 - | GTGGCTGGACAAGAGGT-AATTGATCACTCCTATCTCTGTCT-----------------------------------------CTCTTTCTCT--------------------TTCTGACTCTCTCCCA-CTCTT---TATTTGTAGTGCTCGCTGCCTGTA |
| droGri2 | scaffold_15126:4437367-4437497 + | GTGGCTGGACAAAAGGT-AGGAGAGCACATTGATCTTGAAGAAACTTGCAGAAA---A--CTCTCTAATACTCTCAC---------TCCATTT------------CTCTAACTCTCCCTCTCTCTTATTTAACTT---C-ATTTTAGTGCGCGCTGTTTATA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:3767606-3767703 + | GTGGCTGGACAAAAGGT-TGGGGGACATTGATCTCGGAGA------------------------------------------CGGCGGTTTAACTGATCCATTCTCTCACATCACTT---CCCTCACAGTGCACGGTGCTTGTA |
| droSim1 | chr2L:3722744-3722841 + | GTGGCTGGACAAGAGGT-TGGGGGACATTGATCTCGGAGA------------------------------------------CGGCGGTTTAACTGATCCATTCTCTCACATCACTT---CCCTCACAGTGCGCGGTGCTTGTA |
| droSec1 | super_5:1870685-1870782 + | GTGGCTGGACAAGAGGT-TGGGGGACATTGATCTCTGAGA------------------------------------------CGGCGGTTTAACTGATCCATTCTCTCACATCACTT---CCCTCACAGTGCGCGGTGCTTGTA |
| droYak2 | chr2L:3769291-3769388 + | GTGGCTGGACAAGAGGT-TGGGGGACATTGATCTCGGAGA------------------------------------------CGGCGGTTTAACTGATCCATTCTCTCACATCACTT---CCCTCACAGTGCGCGATGTTTGTA |
| droEre2 | scaffold_4929:3814030-3814127 + | GTGGCTGGACAAGAGGT-TGGGGGACATTGATCTCGGAGA------------------------------------------CGGCGGTTTAACTGATCCAATCTCTCACATCACTT---CCCTCACAGTGCGCGGTGCTTGTA |
| droAna3 | scaffold_12916:5837615-5837739 + | GTGGCTGGACAAGAGGT-GAGATAACACG-TTCT------------AACCAACTGATATGAAACTACGAATGTTTTTATCGCTGTCGATTCGAACGAT-----CTCTCATATAACCGTACCTATTACAGTGCGCGGTGTCTGTA |
| dp4 | chr4_group3:4962038-4962146 - | GTGGCTGGACAAGAGGTGTGTGGGATACTGATTTTAGAGAAAAAAAAAACCATTAACGTG-------------------------AGGCT--------TCG-TTTCTCACATCATTTT-CCCCTCACAGTGCCCGGTGCCTGTA |
| droPer1 | super_1:6454682-6454788 - | GTGGCTGGACAAGAGGTGTGTGGGATACTGATTTTAGAGAAAA--ATAACCATTAACGTG-------------------------AGGC--------TTCG-TTTCTCACATCATTTT-CCCCTCACAGTGCCCGGTGCCTGTA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
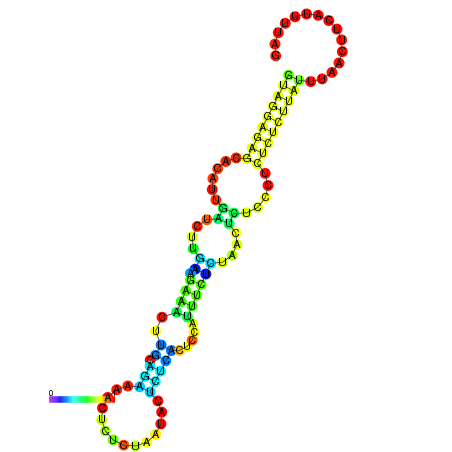
| dG=-19.3, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.7, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
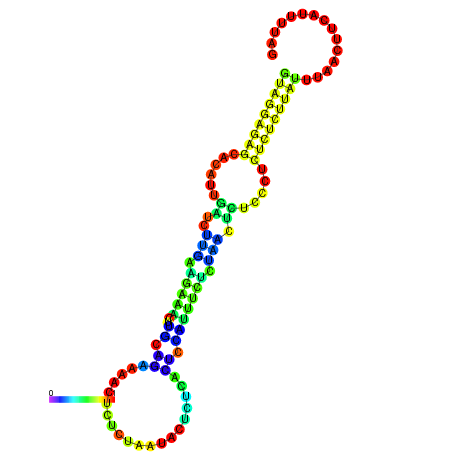
| dG=-19.3, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.7, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
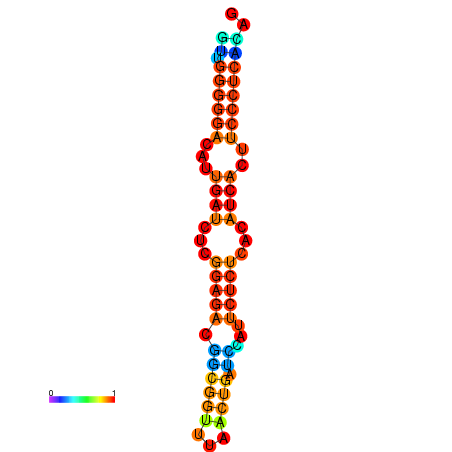 |
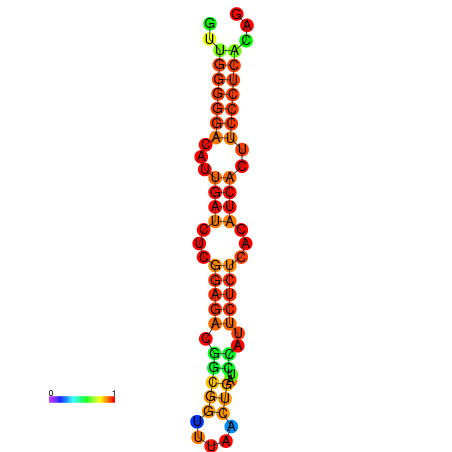 |
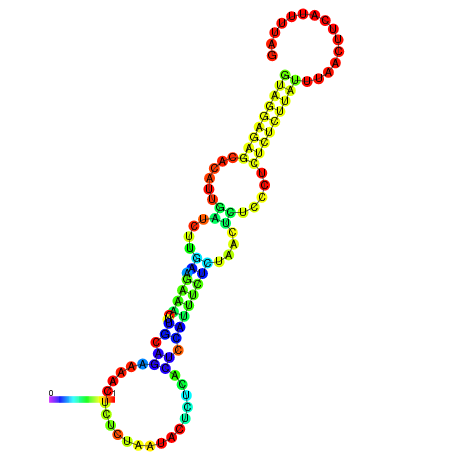
| dG=-18.3, p-value=0.009901 | dG=-18.2, p-value=0.009901 | dG=-18.2, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|---|---|---|
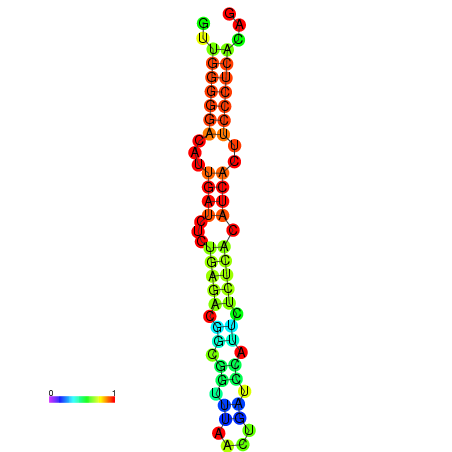 |
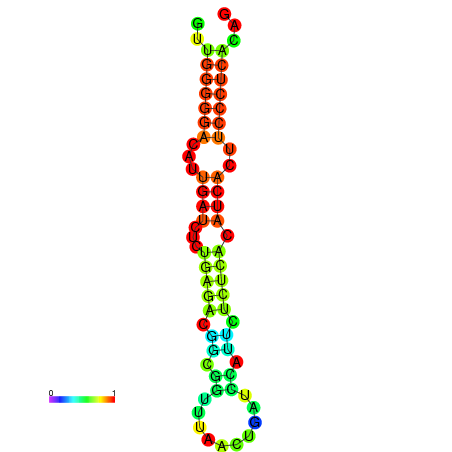 |
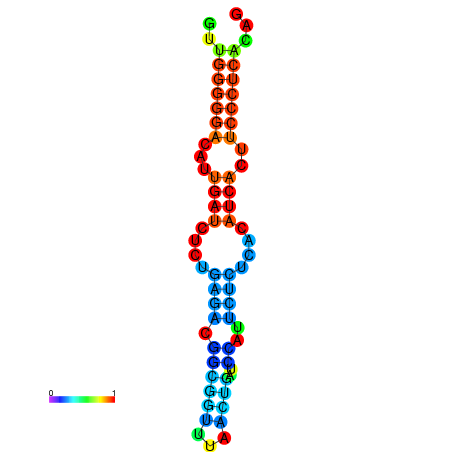 |
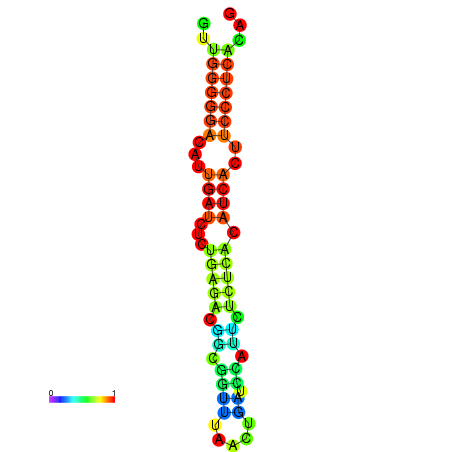 |
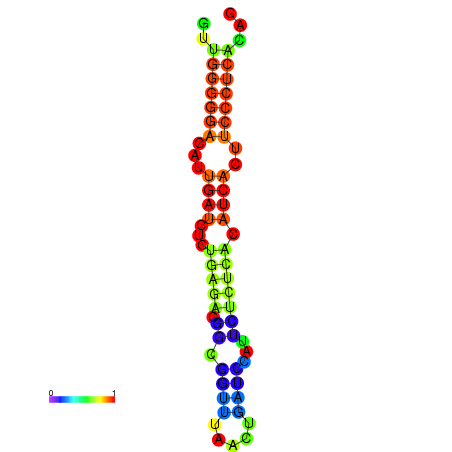 |
| dG=-19.3, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.7, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|---|---|---|
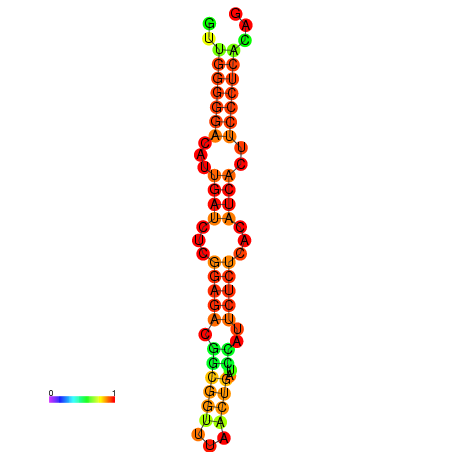 |
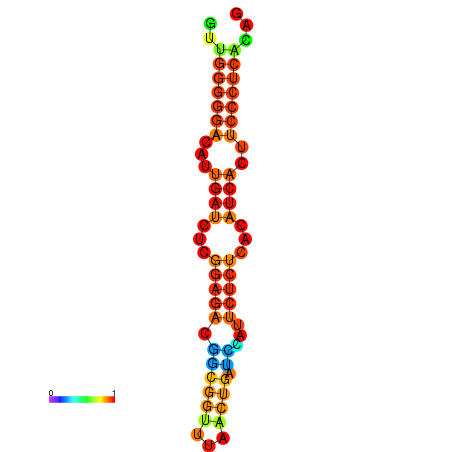 |
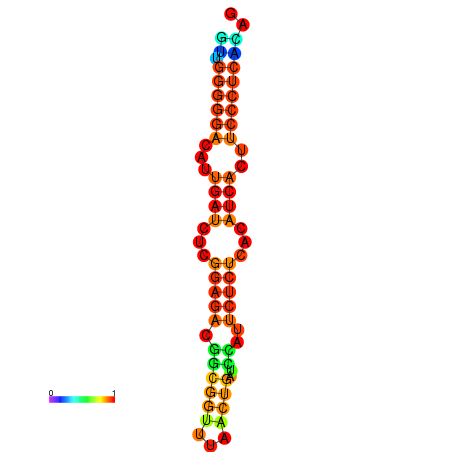 |
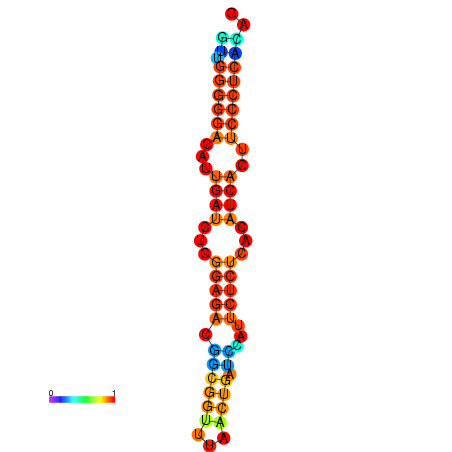 |
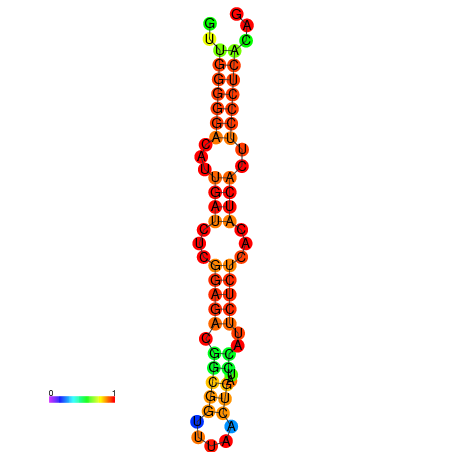 |
| dG=-19.3, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.7, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|---|---|---|
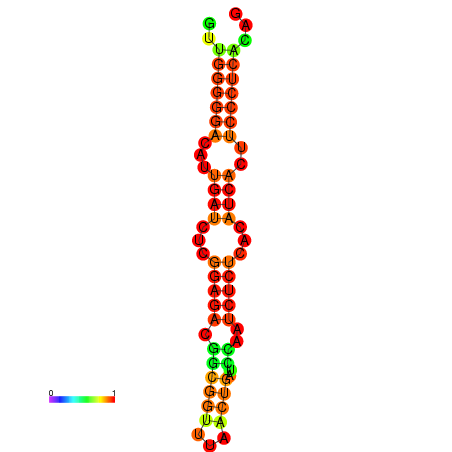 |
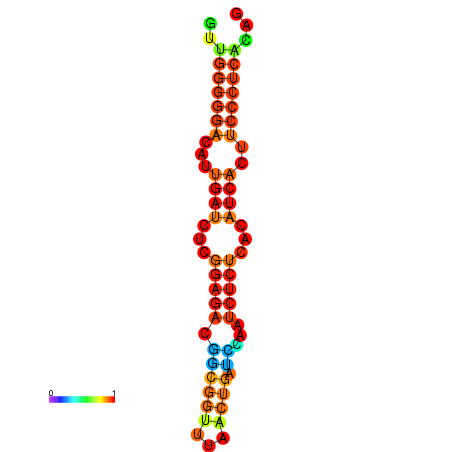 |
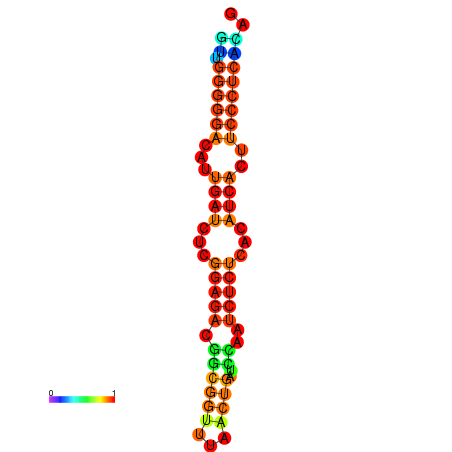 |
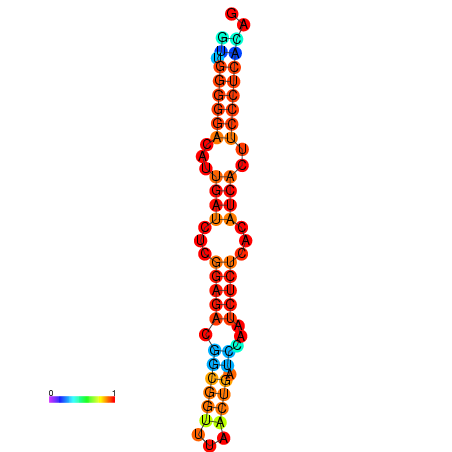 |
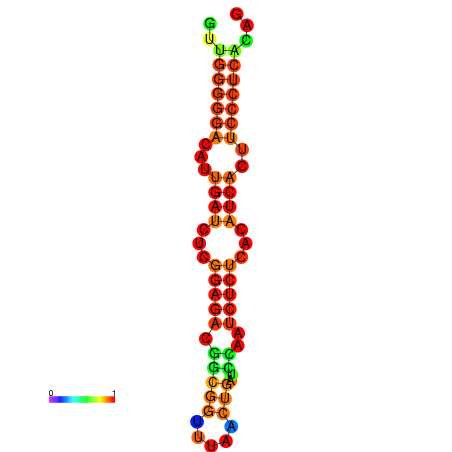 |
| dG=-14.3, p-value=0.009901 | dG=-14.0, p-value=0.009901 | dG=-13.9, p-value=0.009901 | dG=-13.8, p-value=0.009901 | dG=-13.6, p-value=0.009901 |
|---|---|---|---|---|
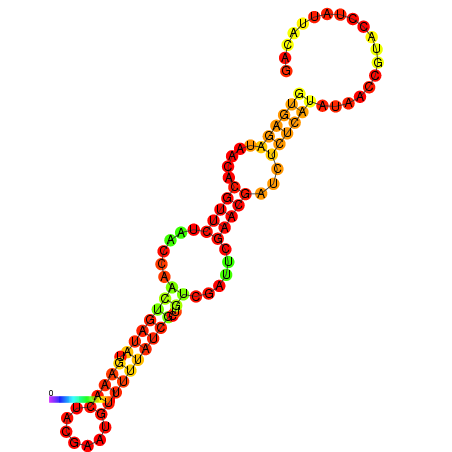 |
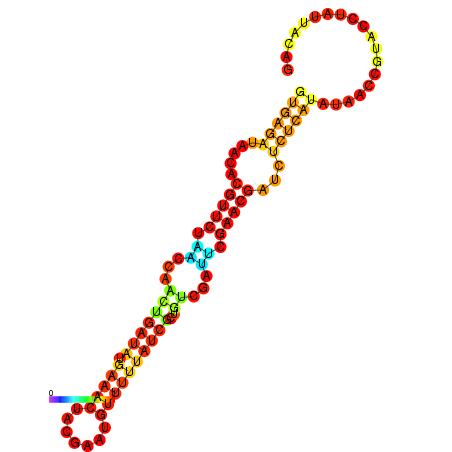 |
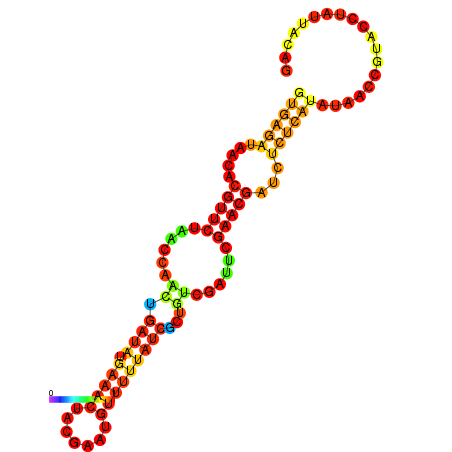 |
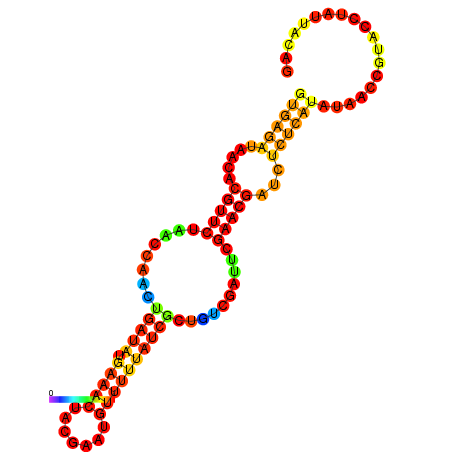 |
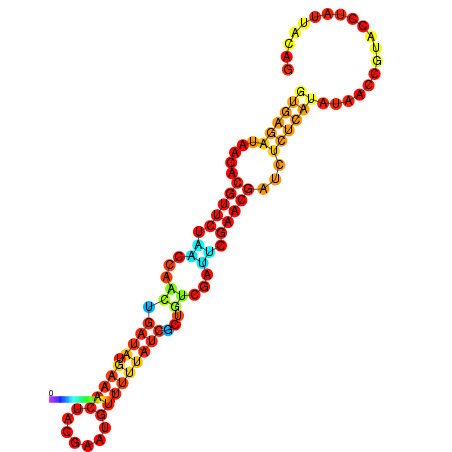 |
| dG=-17.3, p-value=0.009901 | dG=-16.7, p-value=0.009901 | dG=-16.6, p-value=0.009901 | dG=-16.4, p-value=0.009901 | dG=-16.4, p-value=0.009901 |
|---|---|---|---|---|
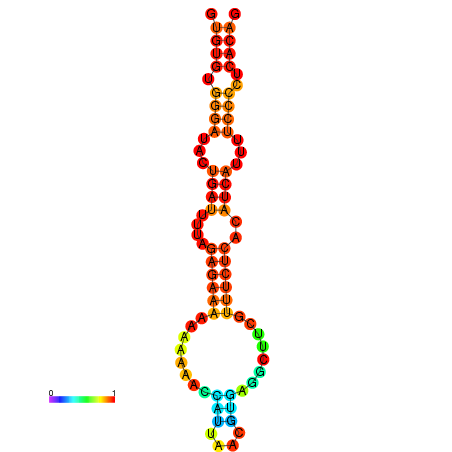 |
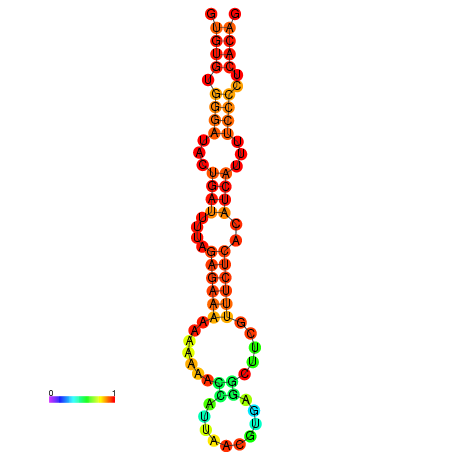 |
 |
 |
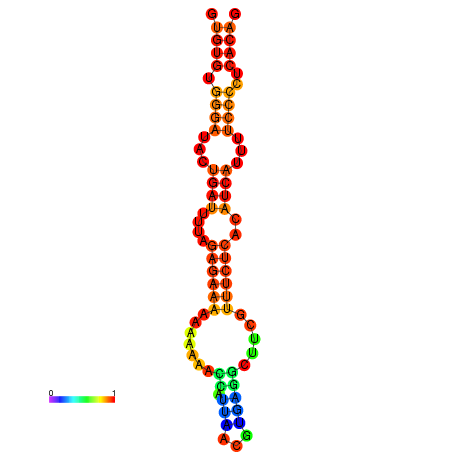 |
| dG=-18.1, p-value=0.009901 | dG=-17.8, p-value=0.009901 | dG=-17.3, p-value=0.009901 |
|---|---|---|
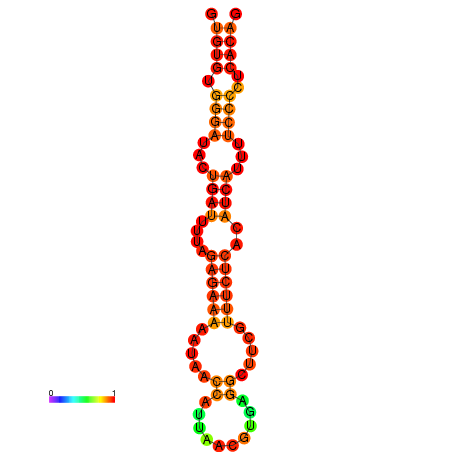 |
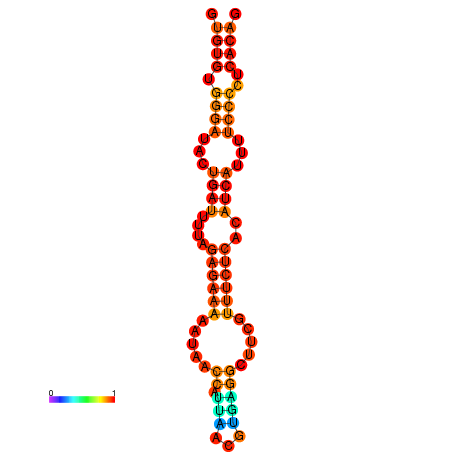 |
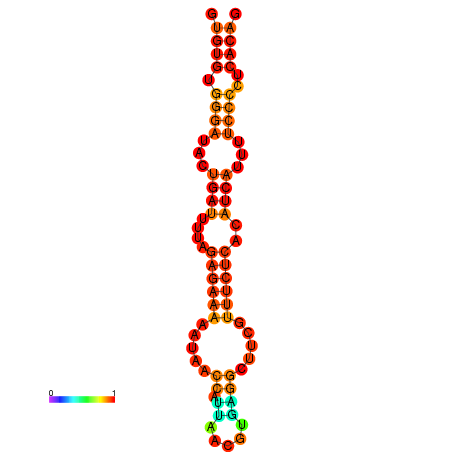 |
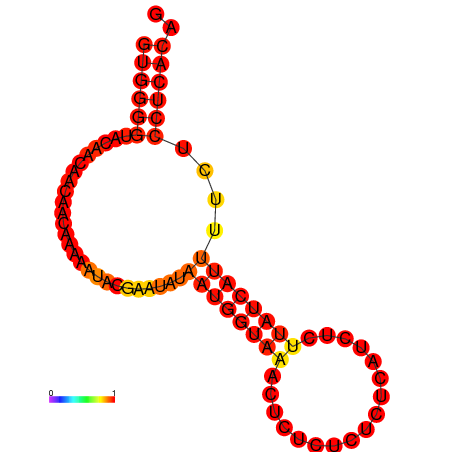
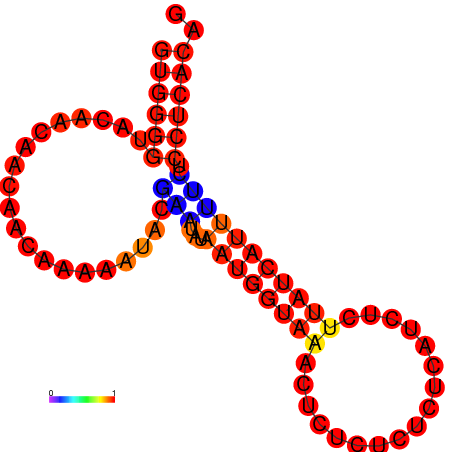
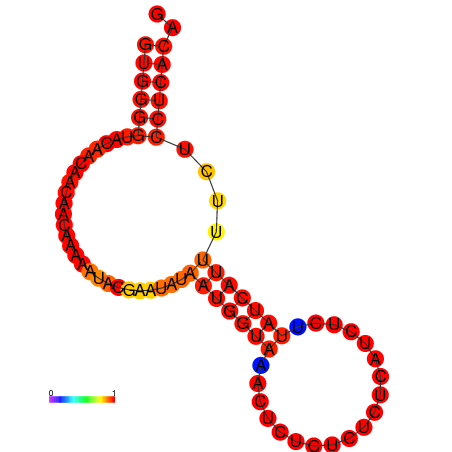
| dG=-8.7, p-value=0.009901 | dG=-7.7, p-value=0.009901 | dG=-7.7, p-value=0.009901 |
|---|---|---|
 |
 |
 |
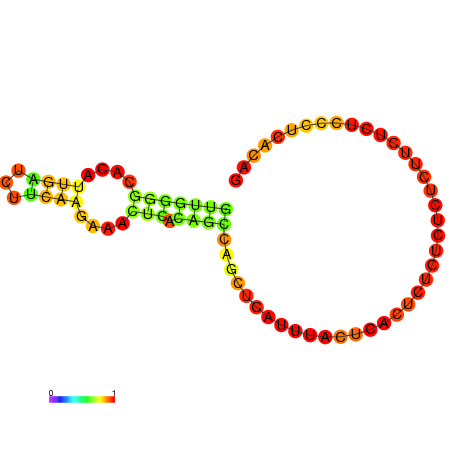
| dG=-6.2, p-value=0.009901 | dG=-5.5, p-value=0.009901 |
|---|---|
 |
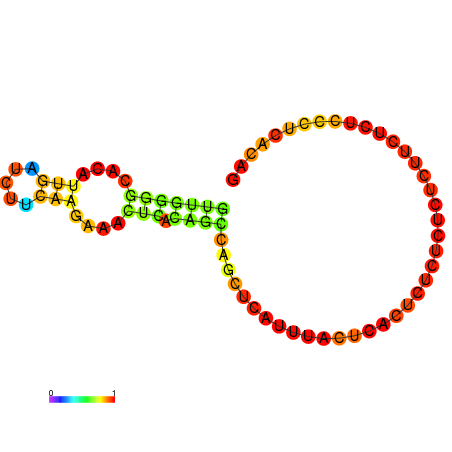 |
| dG=-0.5, p-value=0.009901 | dG=0.0, p-value=0.009901 |
|---|---|
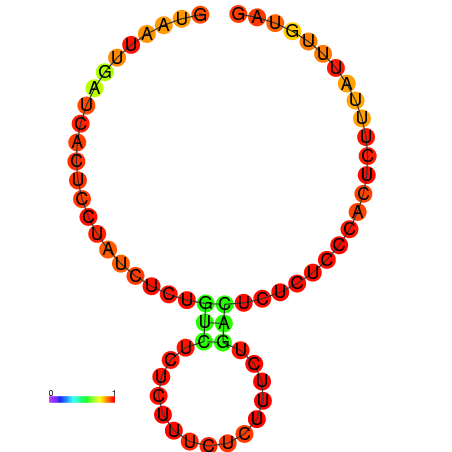 |
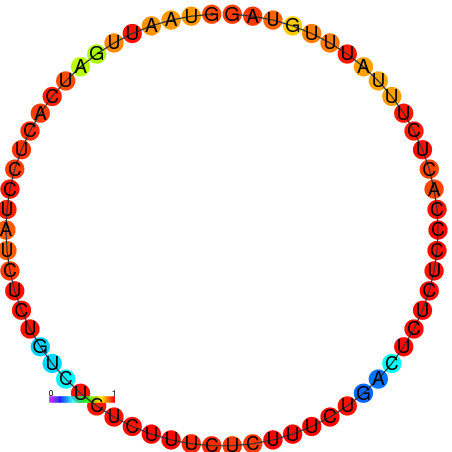 |
| dG=-10.4, p-value=0.009901 | dG=-10.3, p-value=0.009901 | dG=-10.0, p-value=0.009901 | dG=-9.9, p-value=0.009901 | dG=-9.9, p-value=0.009901 |
|---|---|---|---|---|
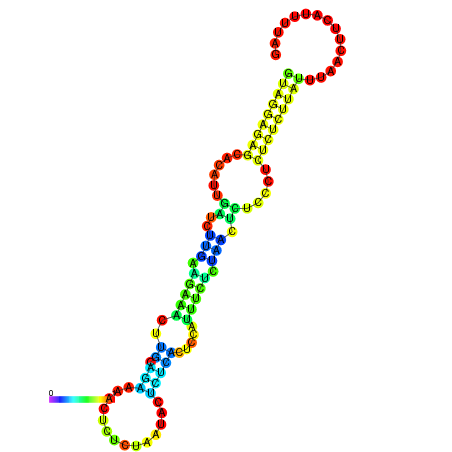 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:46 PM