
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:20484312-20484397 + | CAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droSim1 | chr3R:20323145-20323230 + | GAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droSec1 | super_0:20818704-20818789 + | GAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droYak2 | chr3R:26013202-26013287 - | GAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGTCGTGAGCTGCG |
| droEre2 | scaffold_4820:7574099-7574184 - | GAAACTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droAna3 | scaffold_13340:4690326-4690418 - | CAAGCTACGCCGAAGGTGAGTATAGTGGATGTGGGTGGCTCTTATTTG--------GCCGGTCCTCTCACATCTCCATATTCACAGGCGTCGAGAGCTGCG |
| dp4 | chr2:1751559-1751647 - | GAAGCTACGTAGAAGGTGGGTATCGTGCTTGTGGGTGGCTCTT----G--------GTCAGTCCTCTCACATCTCTATATTCACAGGCGCCGCGAGTTGCG |
| droPer1 | super_7:1904970-1905058 - | GAAGCTACGTAGAAGGTGGGTATCGTGCTTGTGGGTGGCTCTT----G--------GTCAGTCCTCTCACATCTCTATATTCACAGGCGCCGCGAGTTGCG |
| droWil1 | scaffold_181089:9616822-9616911 - | AAAGTTGCGCCGTAGGTGAGTATC-GGGTTGTGATTGGCTCCT----T------GTGGTAGTCCTCTCACATCTCTATATTCACAGGCGTCGCGAGCTGCG |
| droVir3 | scaffold_13047:780932-781019 + | AAAGCTGCGTCGTAGGTGAGTAAT-CAGTTGTGGGTGGCTTTT----T--------GAAAGCCCTCTCACATCTCTTTATTCACAGGCGGCGTGAGTTGCG |
| droMoj3 | scaffold_6540:4775668-4775755 + | CAAGCTGCGGCGCAGGTGAGTAAT-CAGTTGTGGGTGGCTCTA----G--------TGAAGCCCTCTCACATCTCTTTATTCACAGGCGGCGAGAGTTGCG |
| droGri2 | scaffold_14906:5529285-5529380 - | GAAATTGCGGCGCAGGTGAGTAAT-CAGTTGTGGGTGGCTTTT----GTTCTTAGGGAAAGCCCTCTCACATCTCTTTATTCACAGGCGGCGCGAATTGCG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:20484312-20484397 + | CAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droSim1 | chr3R:20323145-20323230 + | GAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droSec1 | super_0:20818704-20818789 + | GAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droYak2 | chr3R:26013202-26013287 - | GAAGCTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGTCGTGAGCTGCG |
| droEre2 | scaffold_4820:7574099-7574184 - | GAAACTGCGCAGAAGGTGGGTATC-TGGATGTGGTTGGCTCT--------------GGCGGTCCTCTCACATTTACATATTCACAGGCGCCGTGAGCTGCG |
| droAna3 | scaffold_13340:4690326-4690418 - | CAAGCTACGCCGAAGGTGAGTATAGTGGATGTGGGTGGCTCTTATTTG--------GCCGGTCCTCTCACATCTCCATATTCACAGGCGTCGAGAGCTGCG |
| dp4 | chr2:1751559-1751647 - | GAAGCTACGTAGAAGGTGGGTATCGTGCTTGTGGGTGGCTCTT----G--------GTCAGTCCTCTCACATCTCTATATTCACAGGCGCCGCGAGTTGCG |
| droPer1 | super_7:1904970-1905058 - | GAAGCTACGTAGAAGGTGGGTATCGTGCTTGTGGGTGGCTCTT----G--------GTCAGTCCTCTCACATCTCTATATTCACAGGCGCCGCGAGTTGCG |
| droWil1 | scaffold_181089:9616822-9616911 - | AAAGTTGCGCCGTAGGTGAGTATC-GGGTTGTGATTGGCTCCT----T------GTGGTAGTCCTCTCACATCTCTATATTCACAGGCGTCGCGAGCTGCG |
| droVir3 | scaffold_13047:780932-781019 + | AAAGCTGCGTCGTAGGTGAGTAAT-CAGTTGTGGGTGGCTTTT----T--------GAAAGCCCTCTCACATCTCTTTATTCACAGGCGGCGTGAGTTGCG |
| droMoj3 | scaffold_6540:4775668-4775755 + | CAAGCTGCGGCGCAGGTGAGTAAT-CAGTTGTGGGTGGCTCTA----G--------TGAAGCCCTCTCACATCTCTTTATTCACAGGCGGCGAGAGTTGCG |
| droGri2 | scaffold_14906:5529285-5529380 - | GAAATTGCGGCGCAGGTGAGTAAT-CAGTTGTGGGTGGCTTTT----GTTCTTAGGGAAAGCCCTCTCACATCTCTTTATTCACAGGCGGCGCGAATTGCG |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
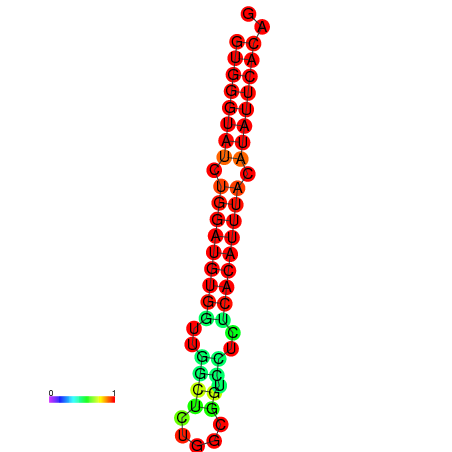
| dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|
 |
 |
 |
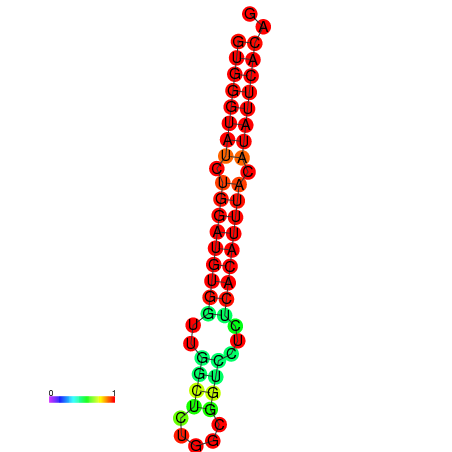
| dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|
 |
 |
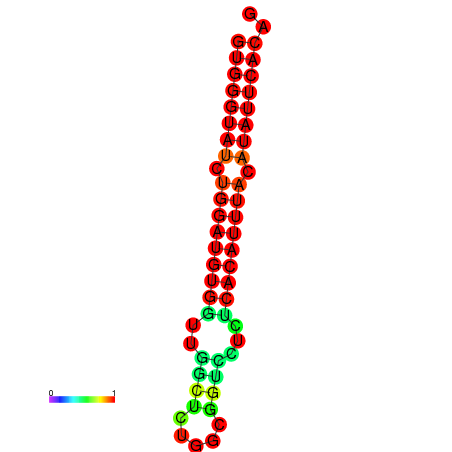 |
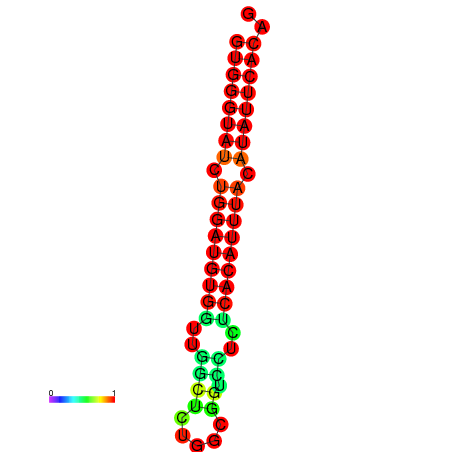
| dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|
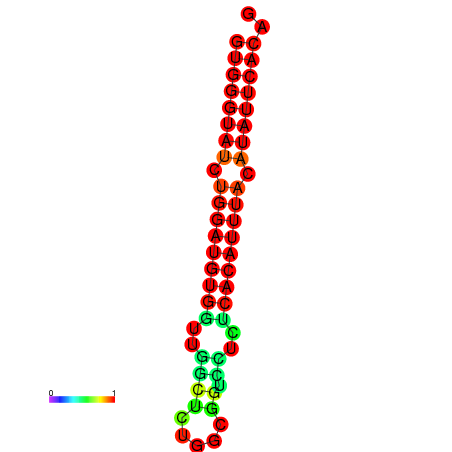 |
 |
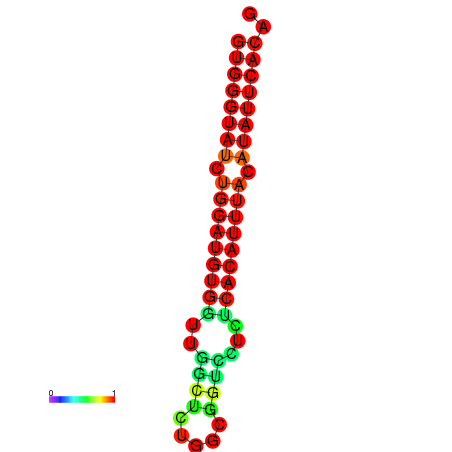 |
| dG=-23.5, p-value=0.009901 | dG=-22.9, p-value=0.009901 |
|---|---|
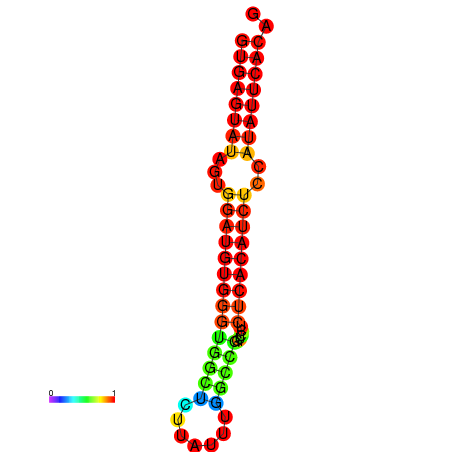 |
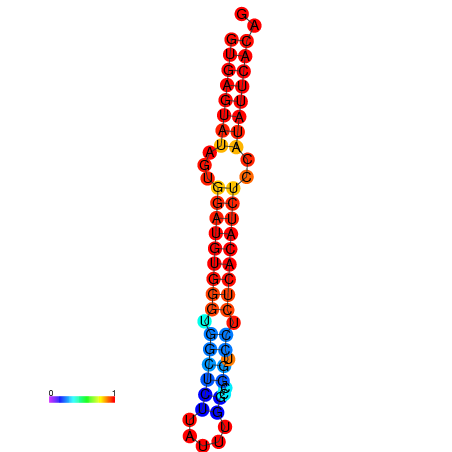 |
| dG=-16.4, p-value=0.009901 | dG=-15.9, p-value=0.009901 | dG=-15.6, p-value=0.009901 |
|---|---|---|
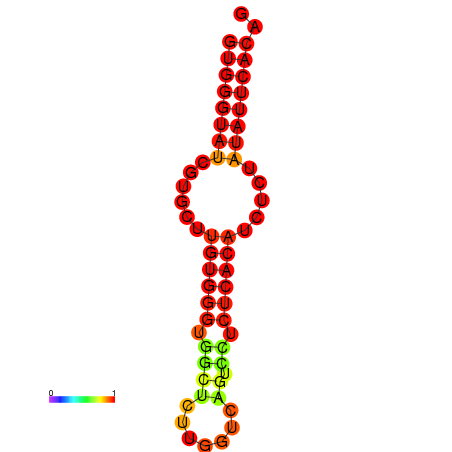 |
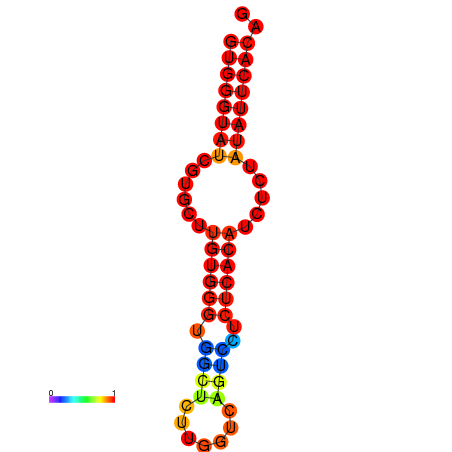 |
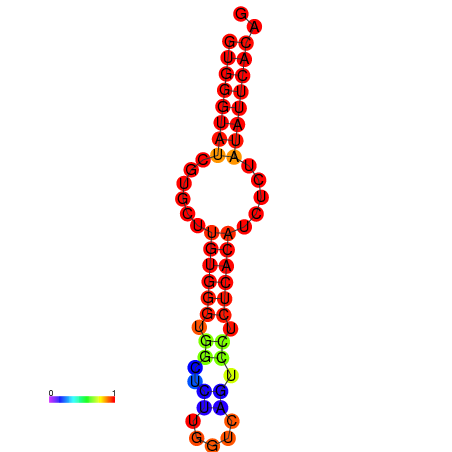 |
| dG=-16.4, p-value=0.009901 | dG=-15.9, p-value=0.009901 | dG=-15.6, p-value=0.009901 |
|---|---|---|
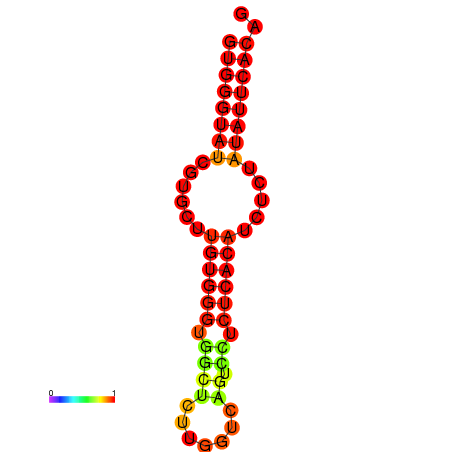 |
 |
 |
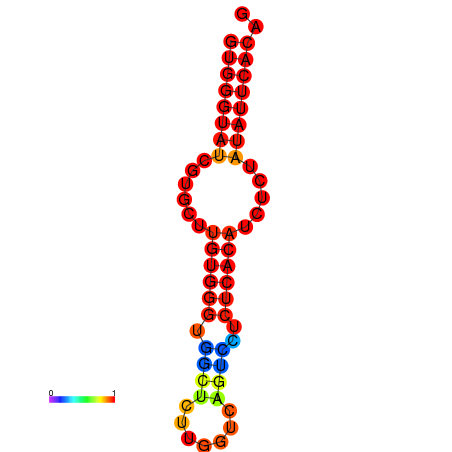
| dG=-25.0, p-value=0.009901 |
|---|
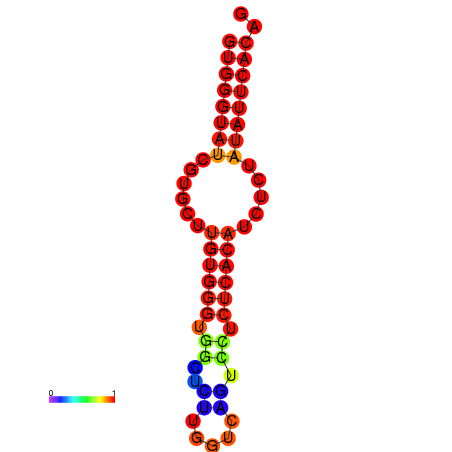
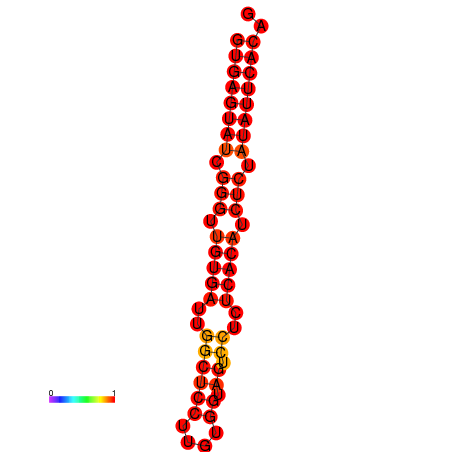 |
| dG=-24.8, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|
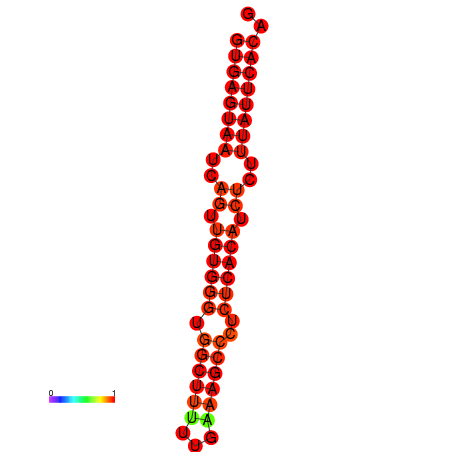 |
 |
| dG=-24.0, p-value=0.009901 | dG=-23.1, p-value=0.009901 |
|---|---|
 |
 |
| dG=-27.4, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|
 |
 |
Generated: 03/07/2013 at 04:44 PM