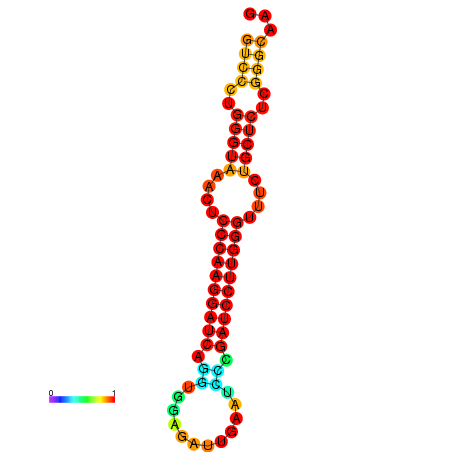
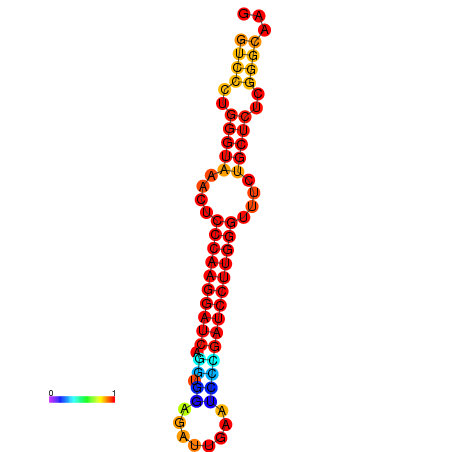
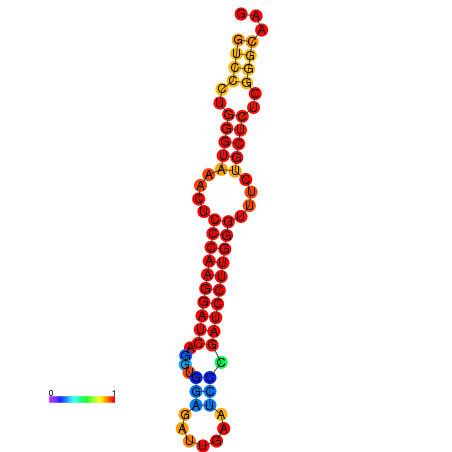
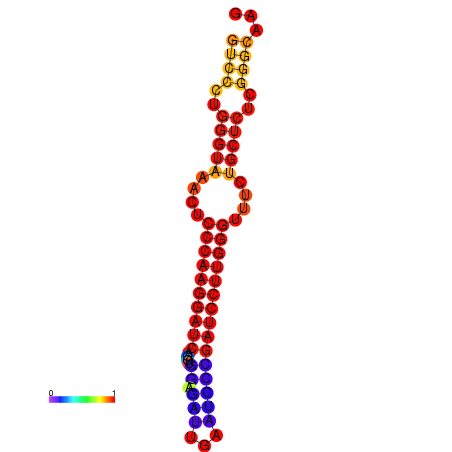
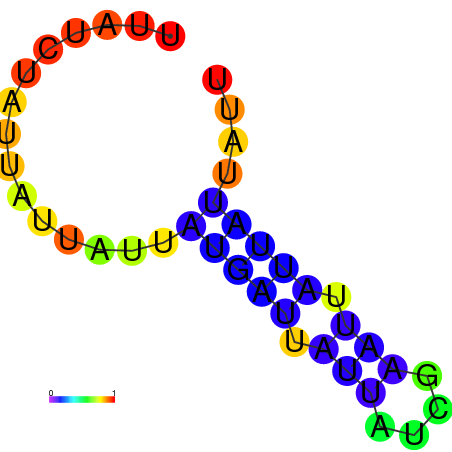
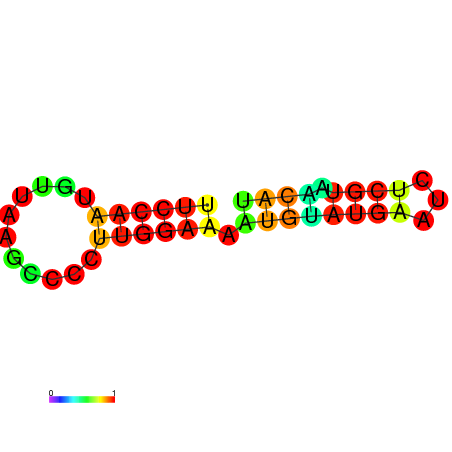
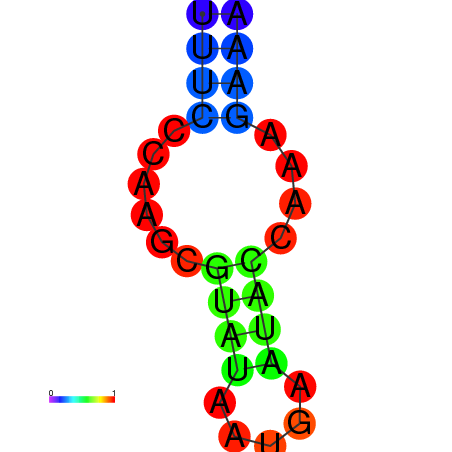
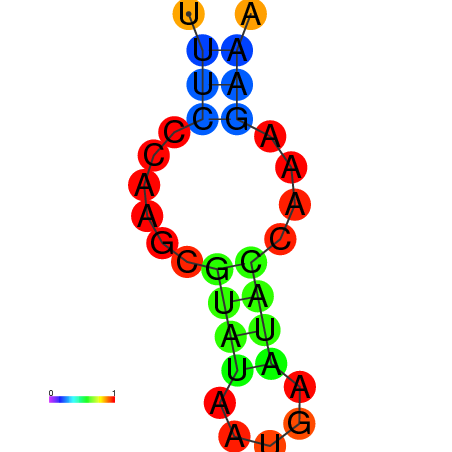
| dm3 |
chr3R:23468173-23468292 - |
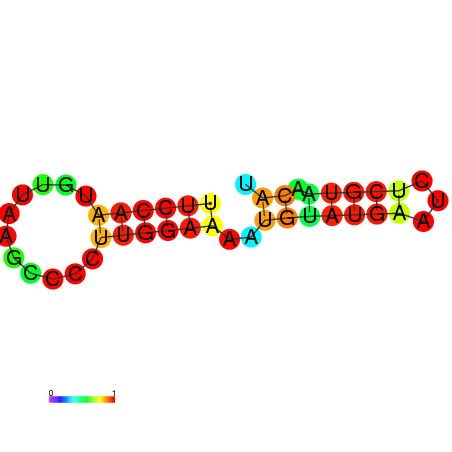
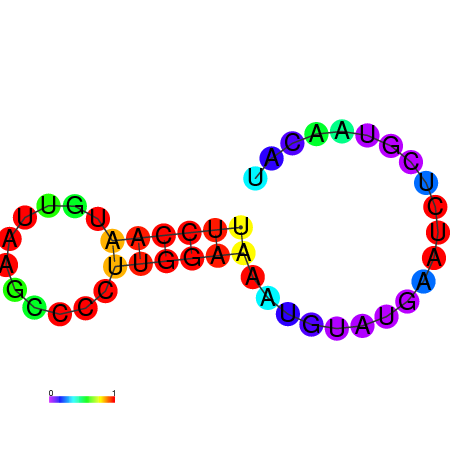
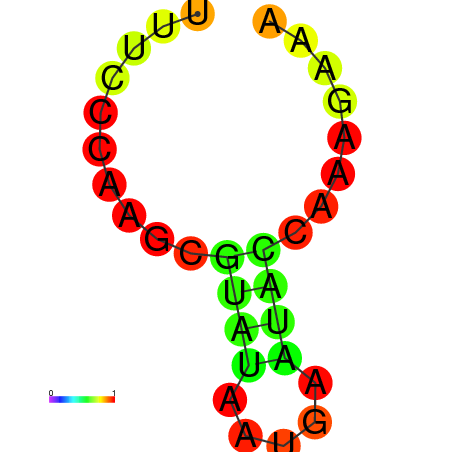
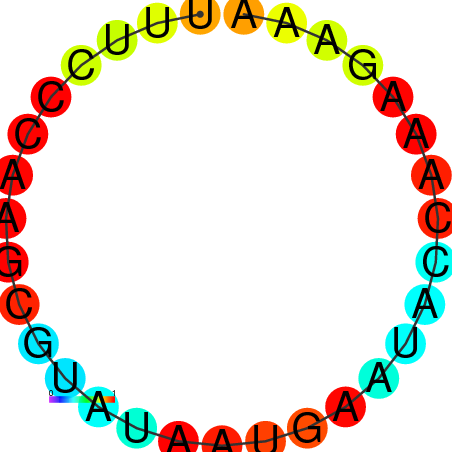
AGCCCGC-----------------------------------------------------------------------------------------GATG-ATGTAAGCTGGC-CTGTCCCTGGGTAAACTCCCAAGGATCAGGTGGAGATTGAATCC---CGATCCTTGGGTTTC------------------TGCTCTCGGGCAAGGTCAGTAGTGCCCGCGAAGGGCCC |
| droSim1 |
chr3R_random:1114763-1114882 - |
AGCCCGC-----------------------------------------------------------------------------------------GATG-ATGTAAGCTGGC-CTGTCCCTGGGTAAACTCCCAAGGATCAGGTGGAGATTGAATCC---CGATCCTTGGGTTTC------------------TGCTCTCGGGCAAGGTCAGCAGTGCCCGCGAAGGGCCC |
| droSec1 |
super_22:111563-111682 - |
AGCCCGC-----------------------------------------------------------------------------------------GATG-ATGTAAGCTGGC-CTGTCCCTGGGTAAACTCCCAAGGATCAGGTGGAGATTGAATCC---CGATCCTTGGGATTC------------------TGCTCTCGGGCAAGGTCAGCAGTGCCCGCGAAGGGCCC |
| droYak2 |
chr3R:23018685-23018798 + |
G------------------------------------------------------------------------------------------------ATG-ATGTAAACTGGC-CTGTCCCTGGGTAAACTCCCAATGATCCGGTGTACATTGAATTC---GGATCCTTGGGTTTG------------------AGCCCAAGGACAAGGACAGTAGTGCCCGCGAAGGGCCC |
| droEre2 |
scaffold_4820:4569064-4569195 + |
AGCCCGCGTAAGGTGTATA-----------------------------------------------------------------------------GATG-ATGTAAACTGGC-CTGTCCCTGGGCAAACTCCCAATGATGCGGTGTACATAGAATCC---CGATCCTTGGGCTTC------------------TGCCCACGGGCAAGGACAGTAGTGCCCGCGAAGGGCCC |
| droAna3 |
scaffold_13340:19221720-19221830 + |
GTGTATA-----------------------------------------------------------------------------------------GATG-ATGTGAGCTGTC-CTGTCCCTGGGAGG----------ATCACCTGTAGTATTAAATTAAATGGAACAAAGGTCTC------------------CCCCTAGGGGCAAGGACAGCATTGGCCACGA--GGCTC |
| dp4 |
chr2:3822386-3822523 - |
AGATGAT-----------------------------------------------------------------------------------------GATACATATAAGCTGATTCTCTCCCAGTGGCGGACACACGGCAGATGGTGGCTTTACCGATC---CGATAATTGAATTTTAACCAAAAGAAAACCACCTGCATTCGGGTGCCGCCCCT-GGGTCTGAG-AGGGCTG |
| droPer1 |
super_7:528167-528304 + |
AGATGAT-----------------------------------------------------------------------------------------GATACATATAAGCTGATTCTCTCCCAGGGGCGGATACACGGCAGATGGTGGCTTTACCGATC---CGATAATTGAATTTTAACCAAAAGAAAACCACCTGCTTTCGGGTGCCGCCCCT-GGGTCTGAG-AGGGCTG |
| droWil1 |
scaffold_181089:3707083-3707253 - |
CAAATGG--AAAGTGTATGGAGGCAAGAGTGTCCAGATTGCCTTGGCCAAATATTGGGATTATGCTCAATACCGATCCCACATAATTGTTAATTCGTTTA-GTATTATCTA-----------------TTATCTATTATTATTAT---------------------------------------GATTATTATCGAATTATTATTATTATTCTT-ATGATTATT-GAGATCC |
| droVir3 |
Unknown |
AGCTGAA-----------------------------------------------------------------------------------------ATTT-GTTTATACC----CTTT-CCAATGTTAAGCCCCTTGGAAAATGTA-----TGAATCT---CGTAACAT-------------------------------------------TCGTAACAAGATGTTCGCCA |
| droMoj3 |
Unknown |
AGCTGAA-----------------------------------------------------------------------------------------ATTG-GTTTATGCTC-----------------TTTCCCAAGCGTATAATGAATAC-------------------------------------------------CAAAGAAAATACCTAACACTCGTAAGAAG--- |
| droGri2 |
Unknown |
AGCTAGC-----------------------------------------------------------------------------------------CA-------------------------------------------------------------------------------------------------------------------------------------- |