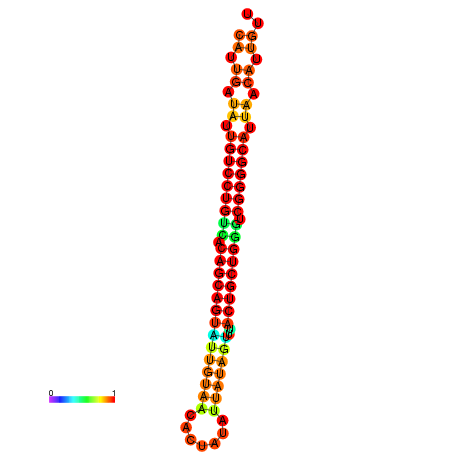
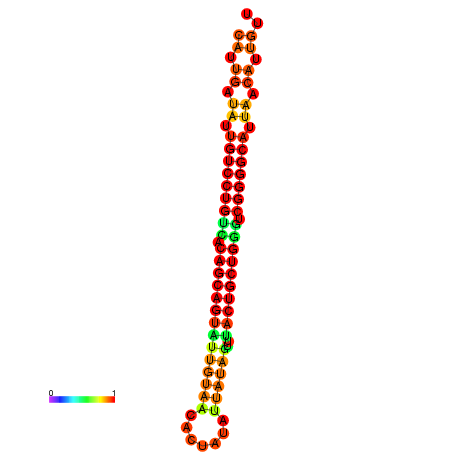
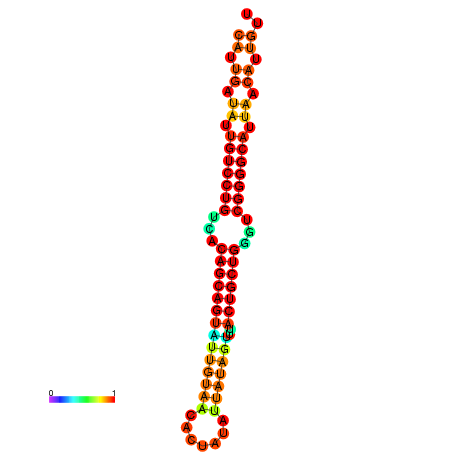
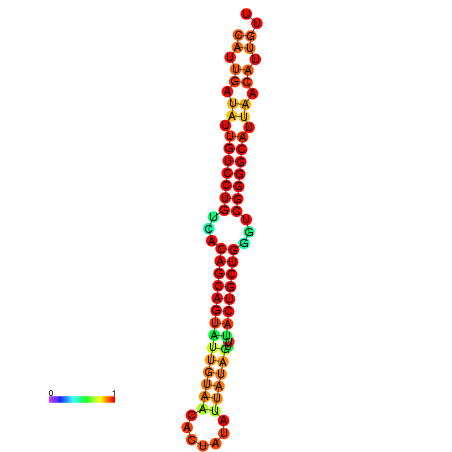
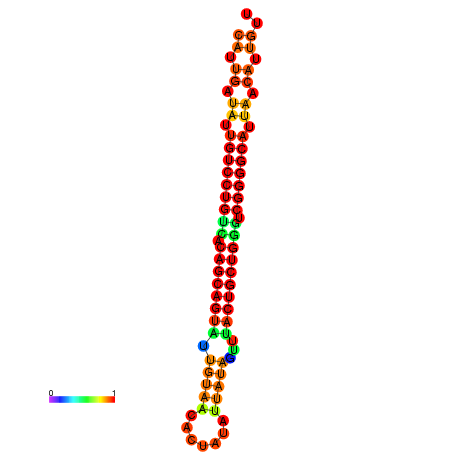
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:21414566-21414695 - | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGTTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGACA-----G- |
| droSim1 | chr3R:21243636-21243765 - | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTTACACTATATT--ATAGTTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGACA-----G- |
| droSec1 | super_13:202504-202633 - | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGTTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGACA-----G- |
| droYak2 | chr3R:25064545-25064667 + | --------GAA----------------TT-------------GG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGCTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGAGG---------------------------------------------------------------------------------CGGCCA-----G- |
| droEre2 | scaffold_4820:6627814-6627943 + | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGCTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGTCA-----G- |
| droAna3 | scaffold_13340:3670416-3670527 - | TA-------------------------------------------GACGCTTGCCGATGATATTGTCCTGTCACAGCAGTATTGTAATTCTATACTATATAGCTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGG----------------------------------------------------------------------------------------A-----G- |
| dp4 | chr2:21158113-21158302 - | TATCGA----ATTACATACGGCGTCGGATCGGAGCAGATGAGAG-GGCGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAAC--TTAATT--ATAGCCTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGCCATTGGCAACA--------AAGAGAAGGAGAAGCTACAAAGCTACAAAGCTACGGAAAAATGCT--------------------------------------------ACA-----G- |
| droPer1 | super_3:3932619-3932791 - | TATCGA----ATTACATACGGCGTCGGATCGGAGCAGATGAGAG-GGCGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAAC--TTAATT--ATAGCCTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGCCATTGGCAACA--------AAGAG-----------------CTACAAAGCTACGGAAAAATGCT--------------------------------------------ACA-----G- |
| droWil1 | scaffold_181130:14638697-14638859 + | TCCATCTAACA----------------AA-------------AG-AGAGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATAGTAAC--ATATTC--ATTTCGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTCTGTGGCAACG--------AGCGGAAAACT--------------------------------CTACTTCTGATGATGATAATAATGATTCAA--------------GTCAA-----A- |
| droVir3 | scaffold_12855:6810247-6810376 - | TATATCGTCG-----------------AT-------------CGTTGCGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATTGTACG--TTAAGC--AATTGGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTCGTTAGCAACCAA------AAAGC---------------------------------------------------------------------------------CAGAAA-----A- |
| droMoj3 | scaffold_6540:25791543-25791693 + | TCTATTAT-------------------------------------AGCGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATTGTAAA--TTAAGC--AATTGGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTCGTTAGCAACC--------AGAAGAAATCTAA--------------------------------------------------------TCGAGTCGACTATGCGGCATACACAGAGG- |
| droGri2 | scaffold_14906:2964690-2964824 - | TATAT---TTA----------------AT-------------CGTTGCGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATTGTTAA--TTAAGC--AATTGGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTTGCTAGCAACTAAAAAGCGAGCAG---------------------------------------------------------------------------------CGAGCA-----GC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:21414566-21414695 - | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGTTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGACA-----G- |
| droSim1 | chr3R:21243636-21243765 - | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTTACACTATATT--ATAGTTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGACA-----G- |
| droSec1 | super_13:202504-202633 - | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGTTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGACA-----G- |
| droYak2 | chr3R:25064545-25064667 + | --------GAA----------------TT-------------GG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGCTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGAGG---------------------------------------------------------------------------------CGGCCA-----G- |
| droEre2 | scaffold_4820:6627814-6627943 + | TATACCATGAA----------------AT-------------CG-GACGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAACACTATATT--ATAGCTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGCGG---------------------------------------------------------------------------------CGGTCA-----G- |
| droAna3 | scaffold_13340:3670416-3670527 - | TA-------------------------------------------GACGCTTGCCGATGATATTGTCCTGTCACAGCAGTATTGTAATTCTATACTATATAGCTTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGTCATTAGCAACG--------AGG----------------------------------------------------------------------------------------A-----G- |
| dp4 | chr2:21158113-21158302 - | TATCGA----ATTACATACGGCGTCGGATCGGAGCAGATGAGAG-GGCGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAAC--TTAATT--ATAGCCTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGCCATTGGCAACA--------AAGAGAAGGAGAAGCTACAAAGCTACAAAGCTACGGAAAAATGCT--------------------------------------------ACA-----G- |
| droPer1 | super_3:3932619-3932791 - | TATCGA----ATTACATACGGCGTCGGATCGGAGCAGATGAGAG-GGCGCTTGCCATTGATATTGTCCTGTCACAGCAGTATTGTAAC--TTAATT--ATAGCCTACTGCTGGGTCGGGGCATTAACATTGTTGAGCGCCATTGGCAACA--------AAGAG-----------------CTACAAAGCTACGGAAAAATGCT--------------------------------------------ACA-----G- |
| droWil1 | scaffold_181130:14638697-14638859 + | TCCATCTAACA----------------AA-------------AG-AGAGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATAGTAAC--ATATTC--ATTTCGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTCTGTGGCAACG--------AGCGGAAAACT--------------------------------CTACTTCTGATGATGATAATAATGATTCAA--------------GTCAA-----A- |
| droVir3 | scaffold_12855:6810247-6810376 - | TATATCGTCG-----------------AT-------------CGTTGCGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATTGTACG--TTAAGC--AATTGGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTCGTTAGCAACCAA------AAAGC---------------------------------------------------------------------------------CAGAAA-----A- |
| droMoj3 | scaffold_6540:25791543-25791693 + | TCTATTAT-------------------------------------AGCGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATTGTAAA--TTAAGC--AATTGGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTCGTTAGCAACC--------AGAAGAAATCTAA--------------------------------------------------------TCGAGTCGACTATGCGGCATACACAGAGG- |
| droGri2 | scaffold_14906:2964690-2964824 - | TATAT---TTA----------------AT-------------CGTTGCGCTTGCCAGTGATATTGTCCTGTCACAGCAGTATTGTTAA--TTAAGC--AATTGGTACTGCTGAGCCGGGGCATTAACATTGTTGAGCGTTGCTAGCAACTAAAAAGCGAGCAG---------------------------------------------------------------------------------CGAGCA-----GC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
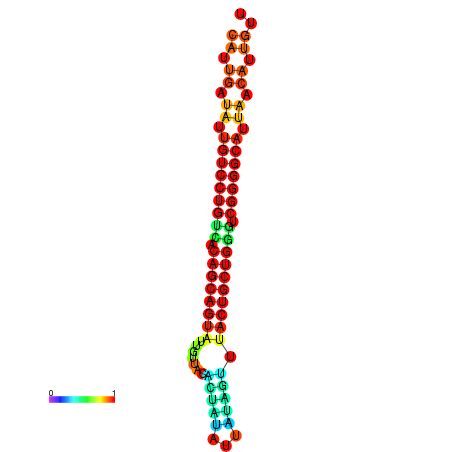
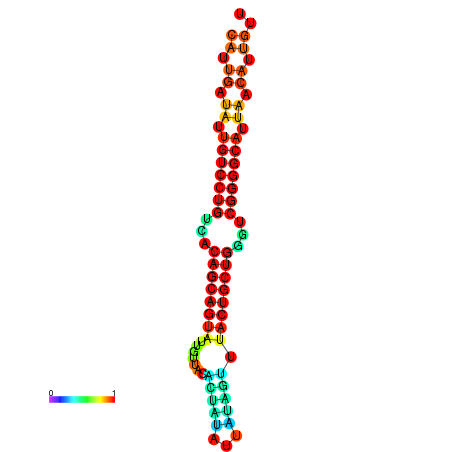
| dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
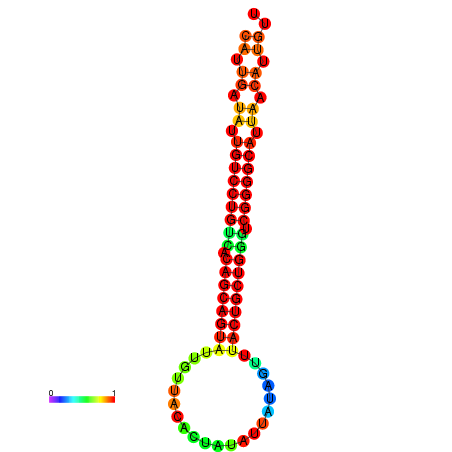 |
 |
 |
| dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
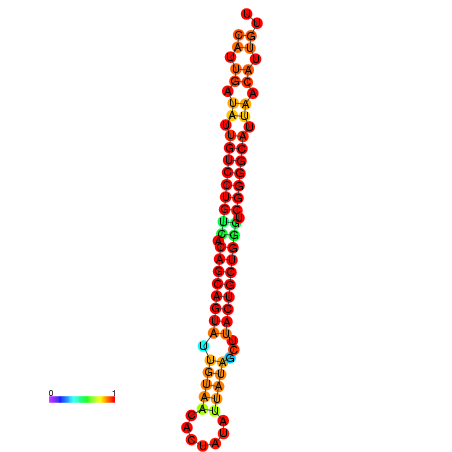
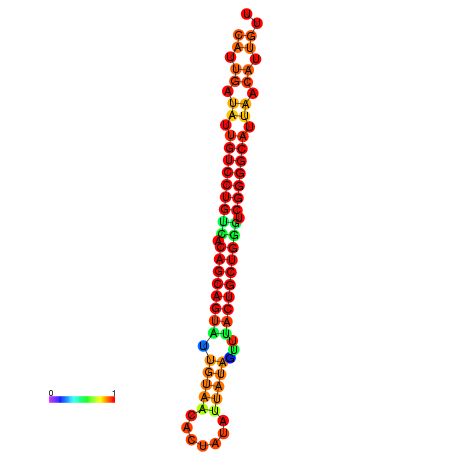 |
| dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|---|---|---|
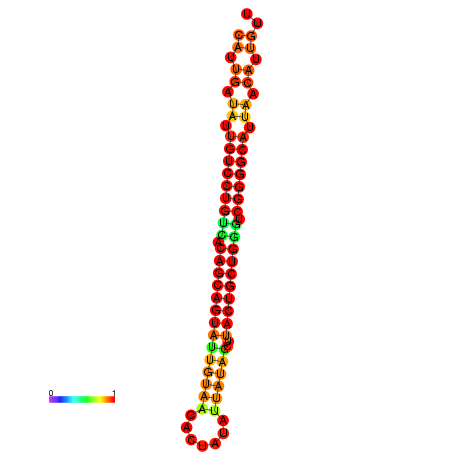 |
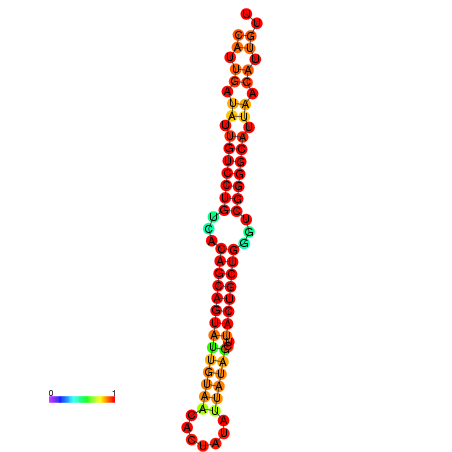 |
 |
 |
 |
| dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
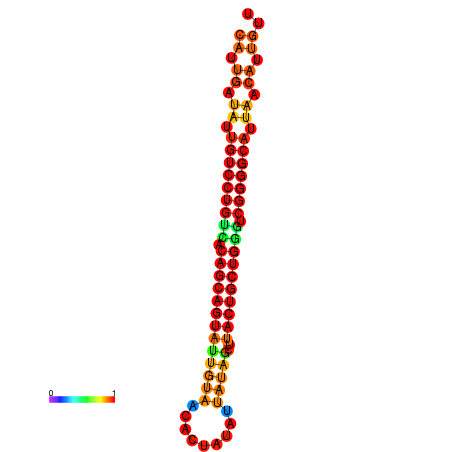 |
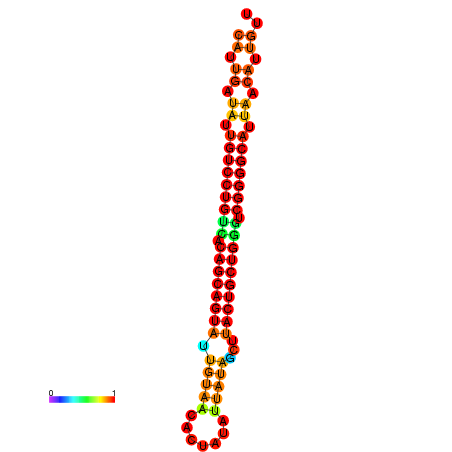
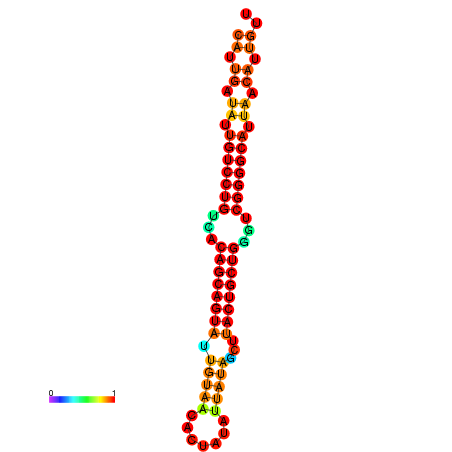
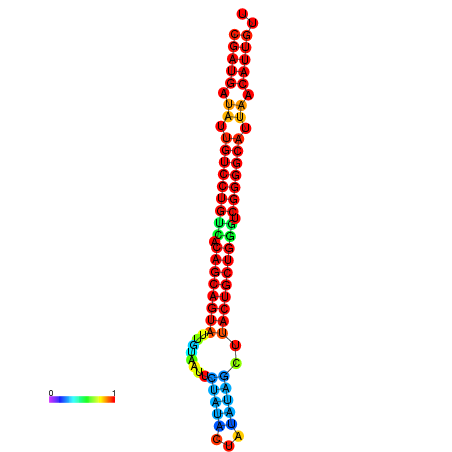
| dG=-28.2, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
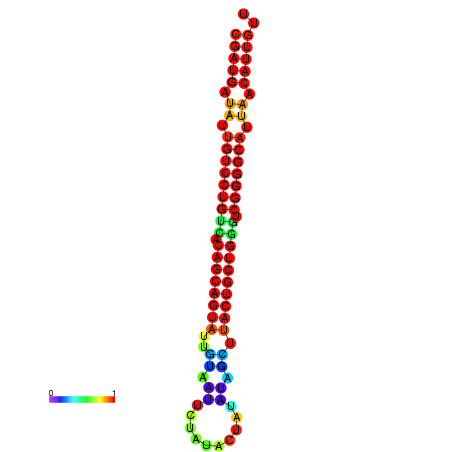 |
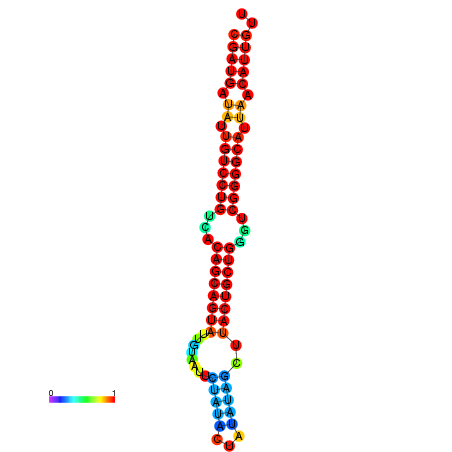
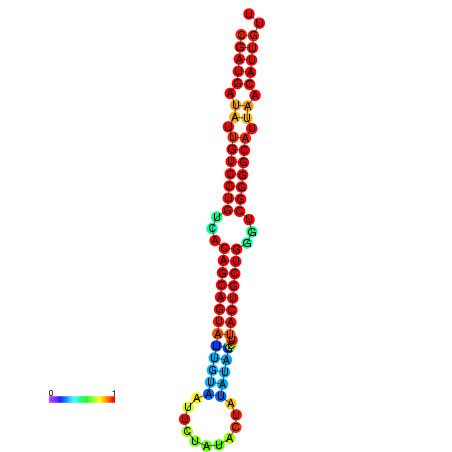
| dG=-26.8, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
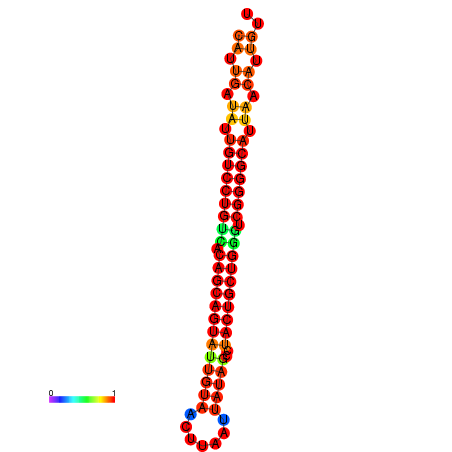 |
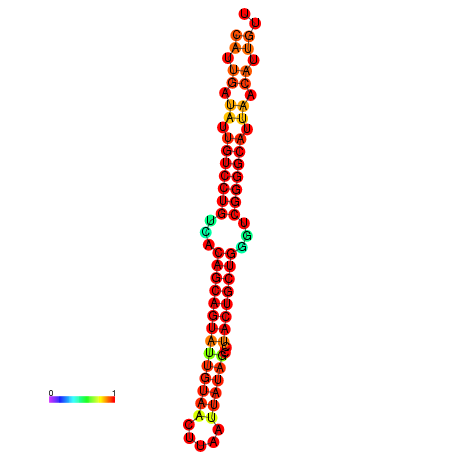
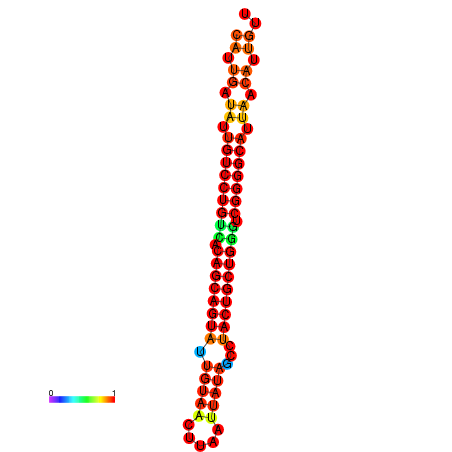
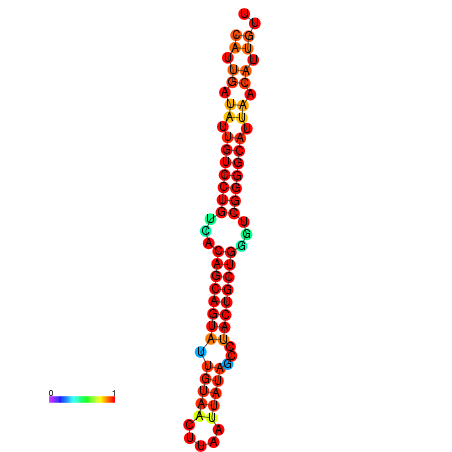
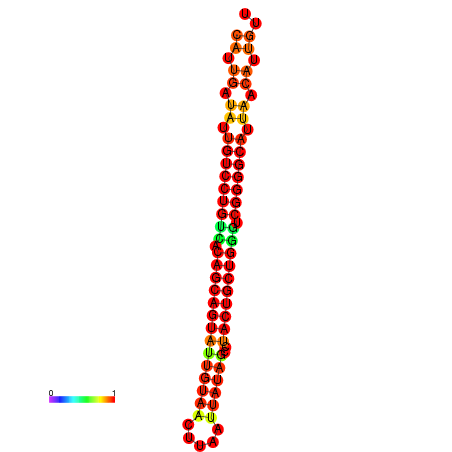
| dG=-26.8, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
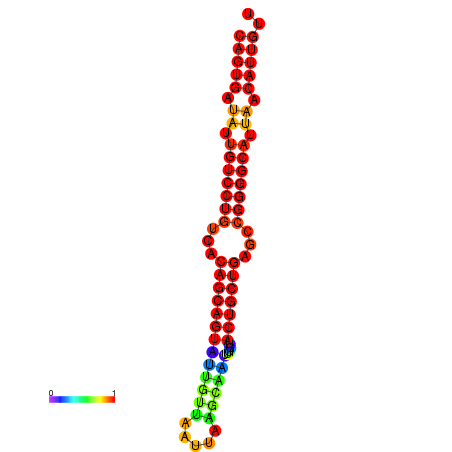
| dG=-27.1, p-value=0.009901 |
|---|
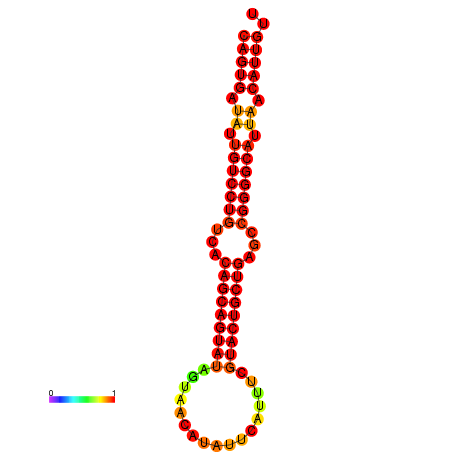 |
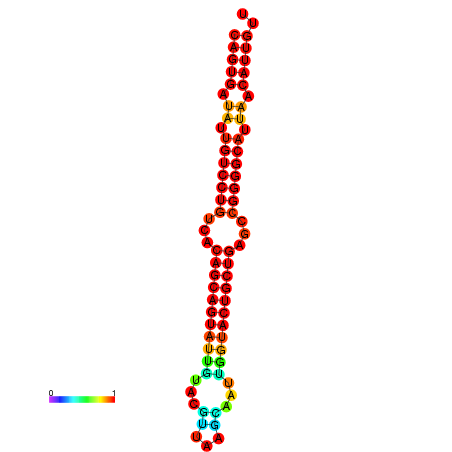
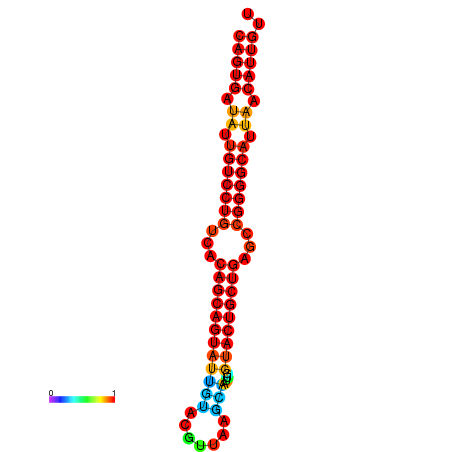
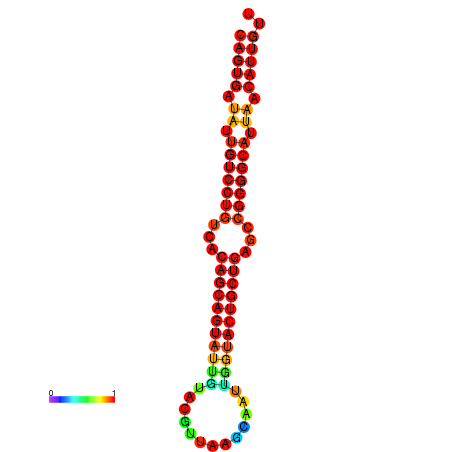
| dG=-28.6, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-28.4, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
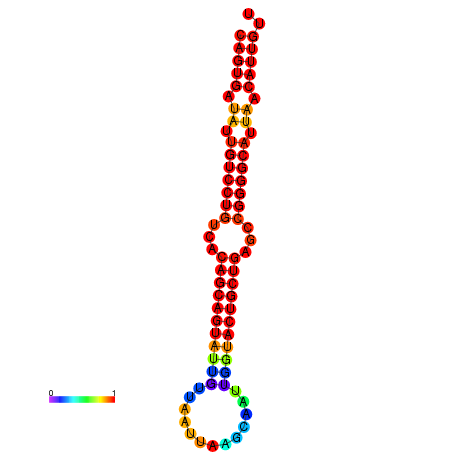
| dG=-29.3, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.6, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:44 PM