
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:18471419-18471548 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droSim1 | chr2L:18160327-18160456 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droSec1 | super_7:2092721-2092850 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droYak2 | chr2R:4988205-4988334 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droEre2 | scaffold_4845:5800004-5800133 - | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droAna3 | scaffold_12916:2378756-2378885 + | ATACTCACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTTTATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATCGAA |
| dp4 | chr4_group3:3634738-3634867 + | ATACTTATATATGGACCATTAACAGAAACCCGTAATTCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------TT-ATCACA |
| droPer1 | super_1:5115309-5115438 + | ATACTTATATATGGACCATTAACAGAAACCCGTAATTCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------TT-ATCACA |
| droWil1 | scaffold_180708:11622810-11622939 + | ATAGTCACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGGCCAACAAAAT---------------------CGAAT--CG |
| droVir3 | scaffold_12963:18449830-18449980 + | ATACTCAAATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAGACATATAAAATATATTAACAAAA-ATGACA |
| droMoj3 | scaffold_6500:6185704-6185833 - | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACTTGGTTTTTGG-CAACAAA-G---------------------CA-AAGCAA |
| droGri2 | scaffold_15252:6427650-6427777 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTTTATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAA-----------------------G-ACTACA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:18471419-18471548 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droSim1 | chr2L:18160327-18160456 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droSec1 | super_7:2092721-2092850 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droYak2 | chr2R:4988205-4988334 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droEre2 | scaffold_4845:5800004-5800133 - | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATGACA |
| droAna3 | scaffold_12916:2378756-2378885 + | ATACTCACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTTTATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------CA-ATCGAA |
| dp4 | chr4_group3:3634738-3634867 + | ATACTTATATATGGACCATTAACAGAAACCCGTAATTCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------TT-ATCACA |
| droPer1 | super_1:5115309-5115438 + | ATACTTATATATGGACCATTAACAGAAACCCGTAATTCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAT---------------------TT-ATCACA |
| droWil1 | scaffold_180708:11622810-11622939 + | ATAGTCACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGGCCAACAAAAT---------------------CGAAT--CG |
| droVir3 | scaffold_12963:18449830-18449980 + | ATACTCAAATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAAGACATATAAAATATATTAACAAAA-ATGACA |
| droMoj3 | scaffold_6500:6185704-6185833 - | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTATATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACTTGGTTTTTGG-CAACAAA-G---------------------CA-AAGCAA |
| droGri2 | scaffold_15252:6427650-6427777 + | ATACTTACATATGGACCATTAACAGAAACCCGTAAATCCGAACTTGTGCTGTTTTTTATCTGTTACAAGACCGGCATTATGGGAGTCTGTCAATGCAAACAACT-GGTTTTTGG-CAACAAAA-----------------------G-ACTACA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
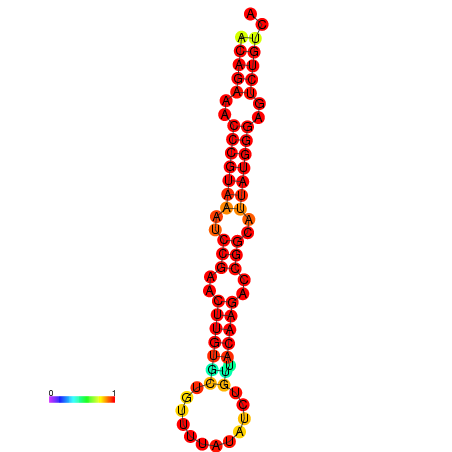
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
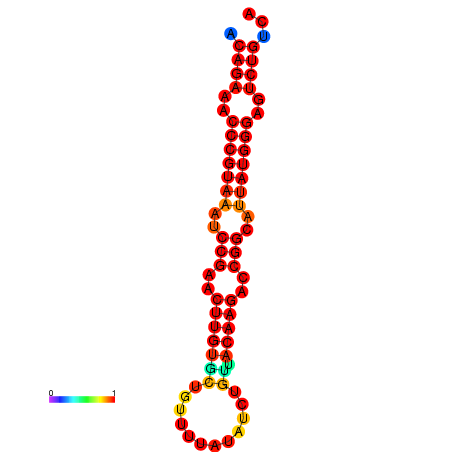
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
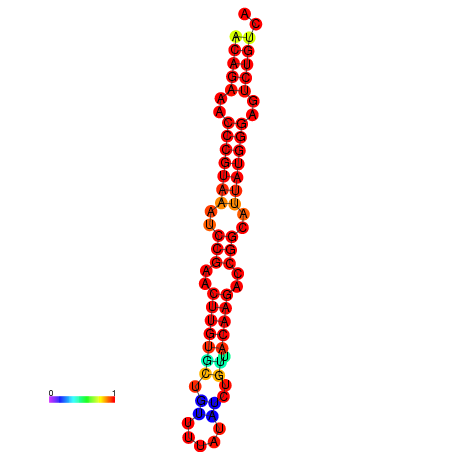 |
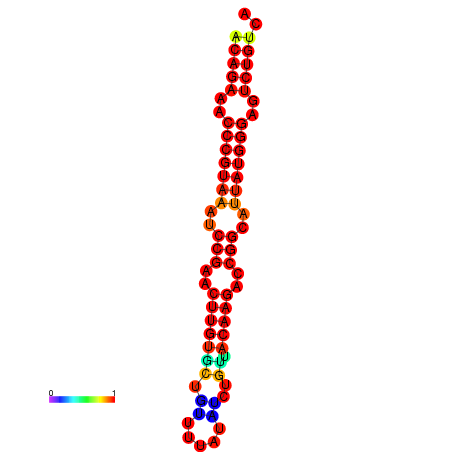
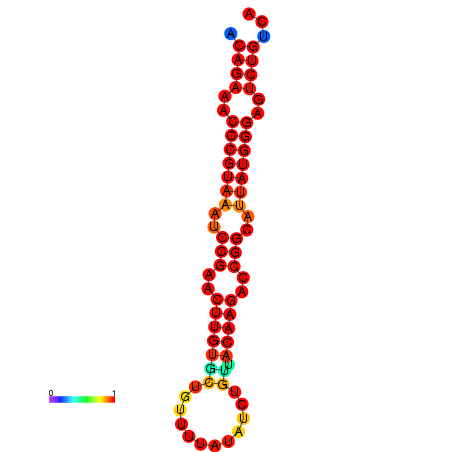
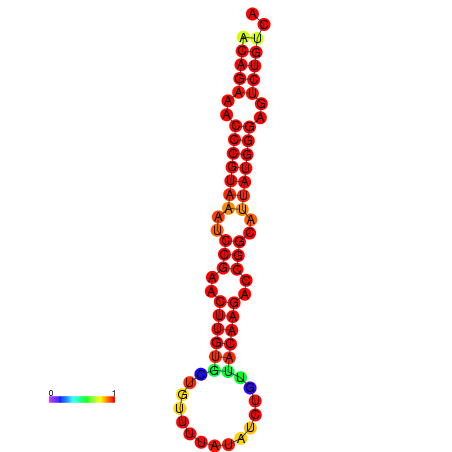
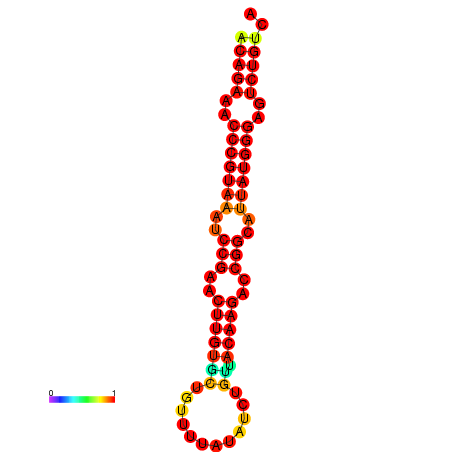
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
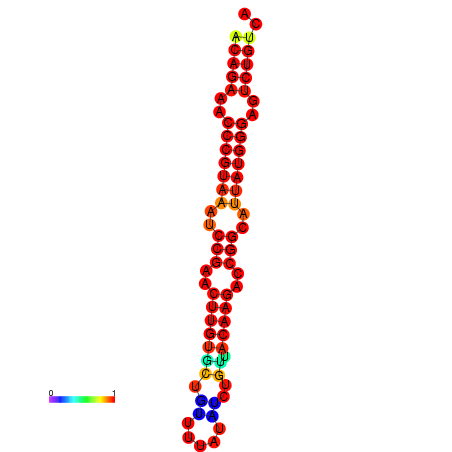 |
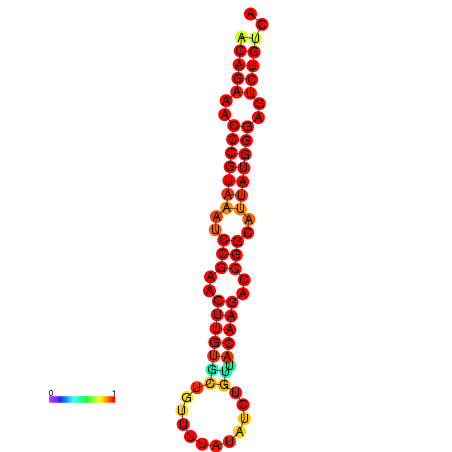
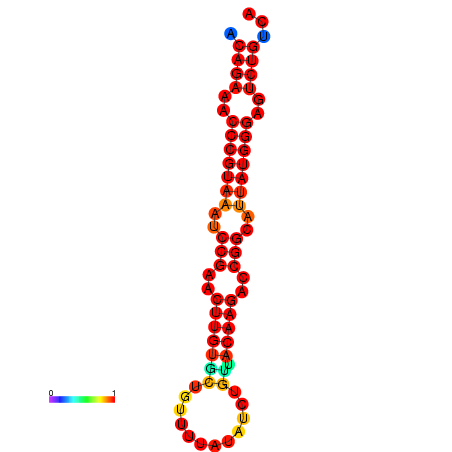
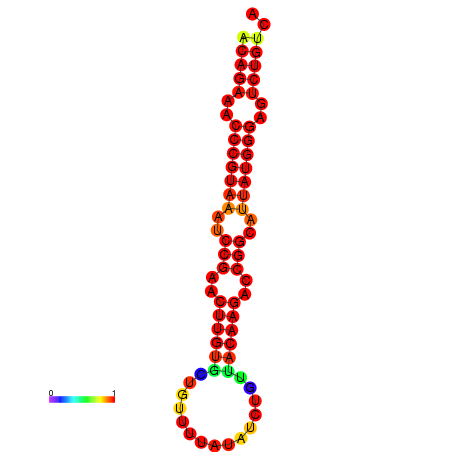
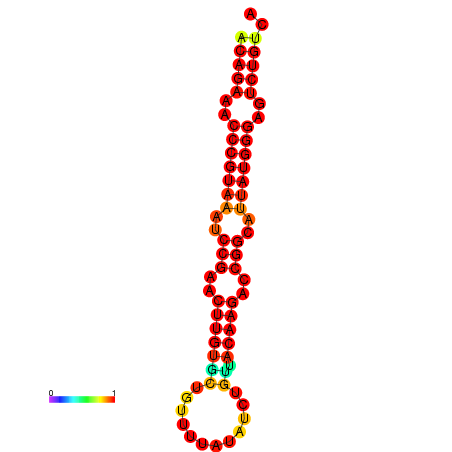
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
 |
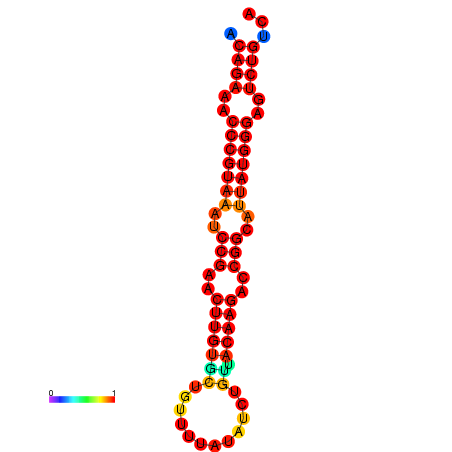 |
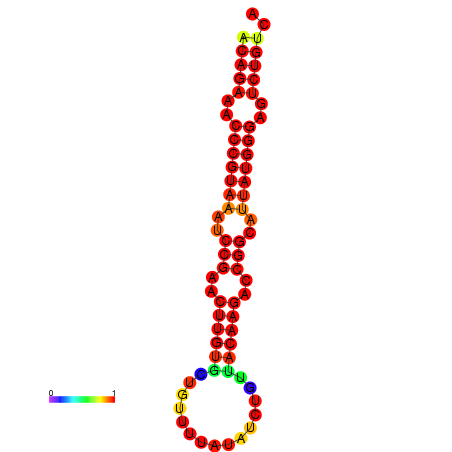 |
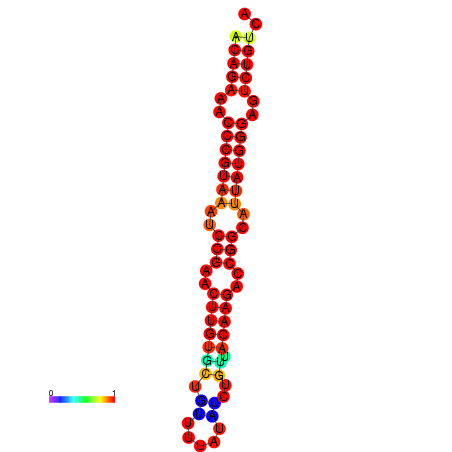 |
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
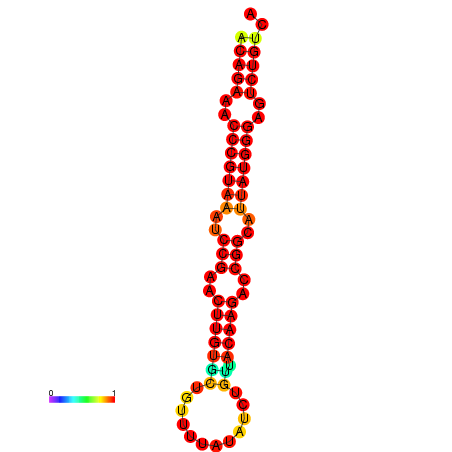 |
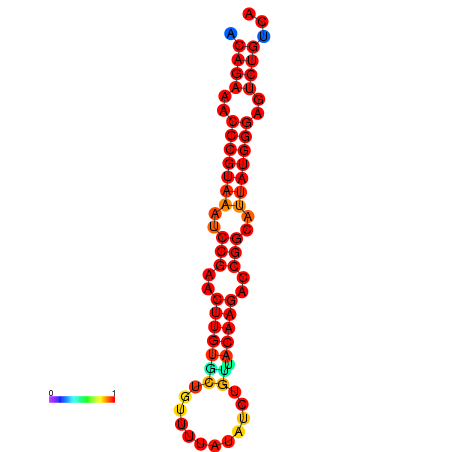 |
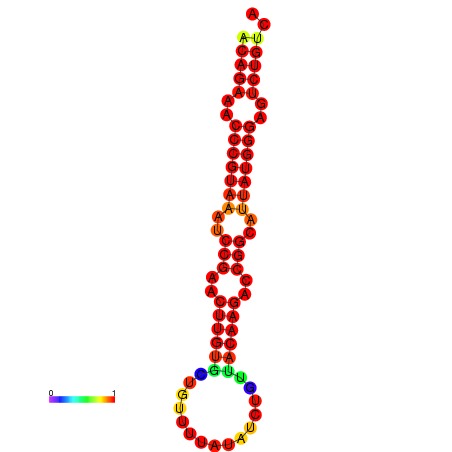 |
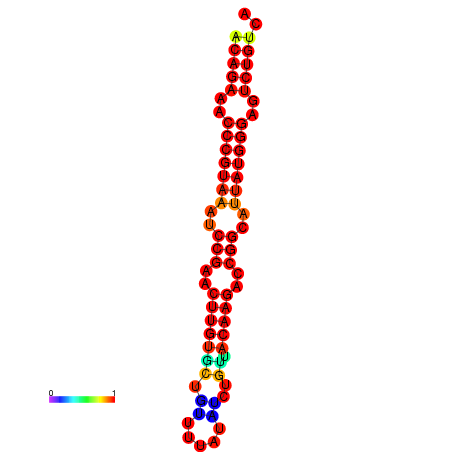 |
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
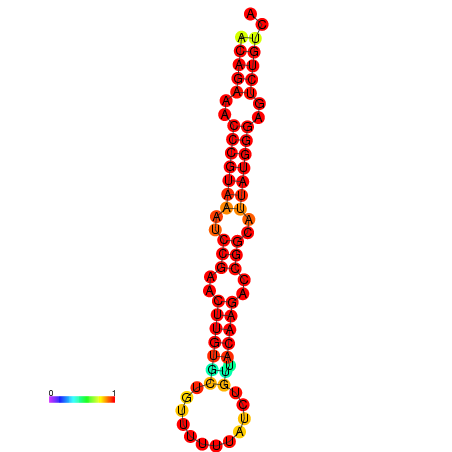 |
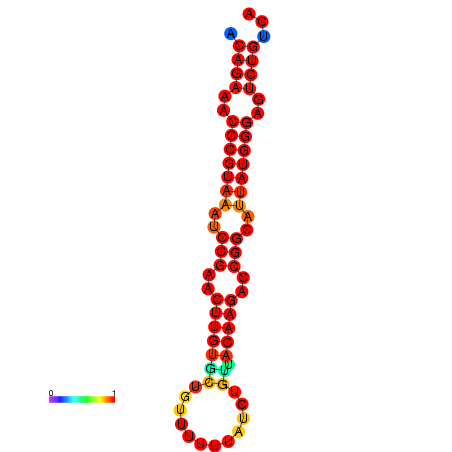 |
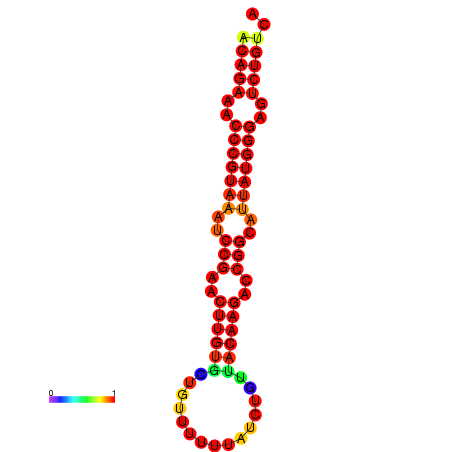 |
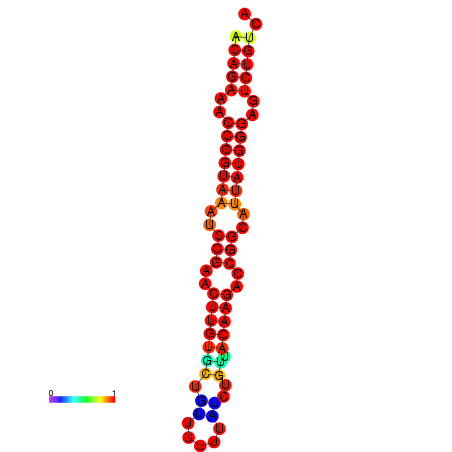 |
| dG=-25.6, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|
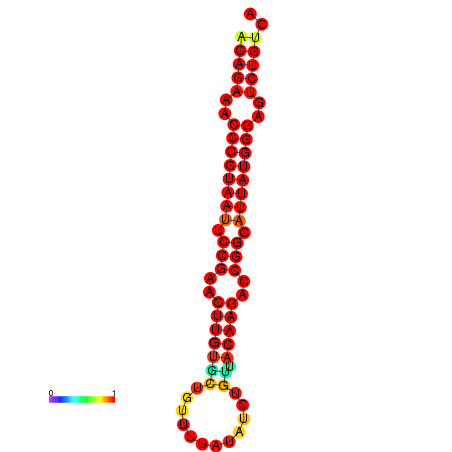 |
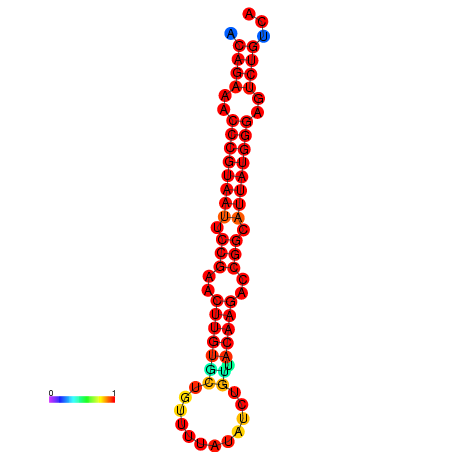 |
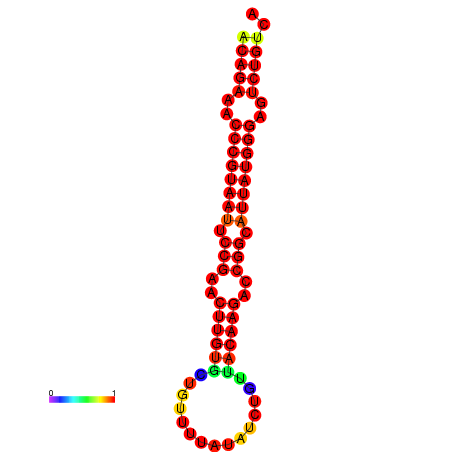 |
 |
| dG=-25.6, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|
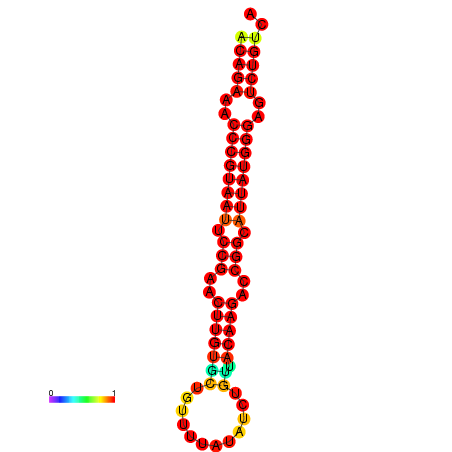 |
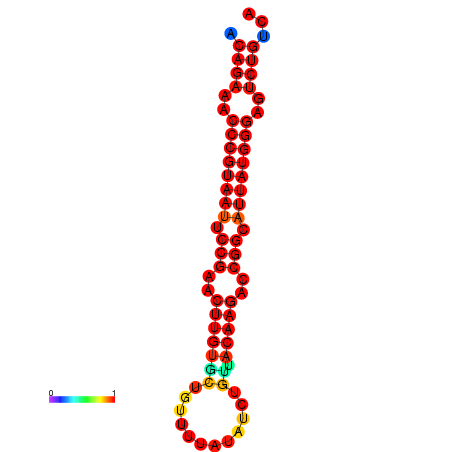 |
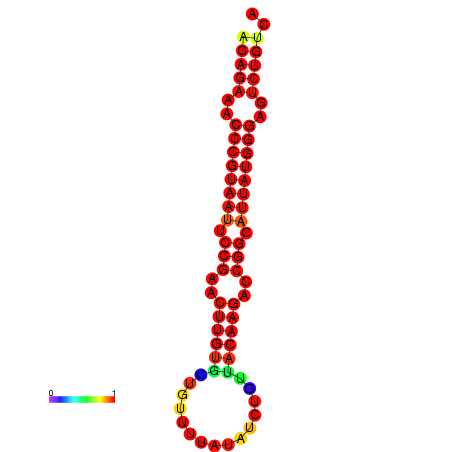 |
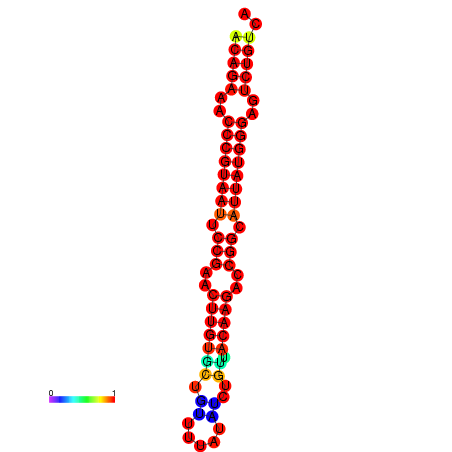 |
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
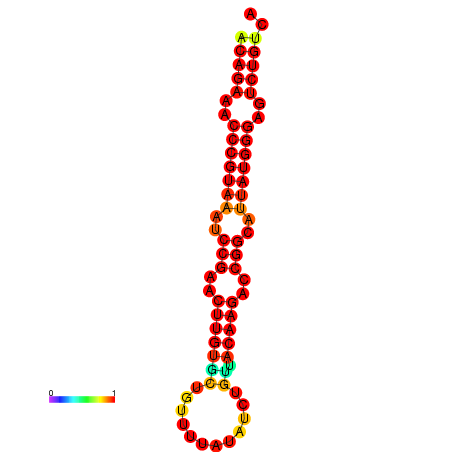 |
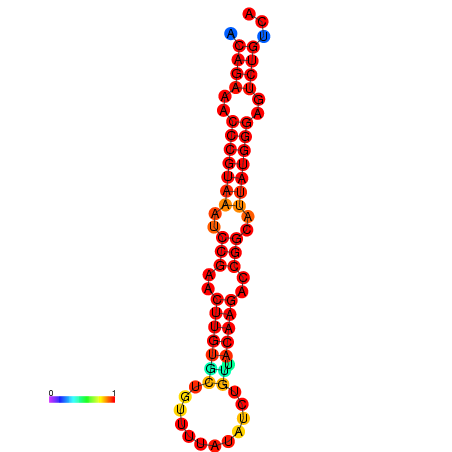 |
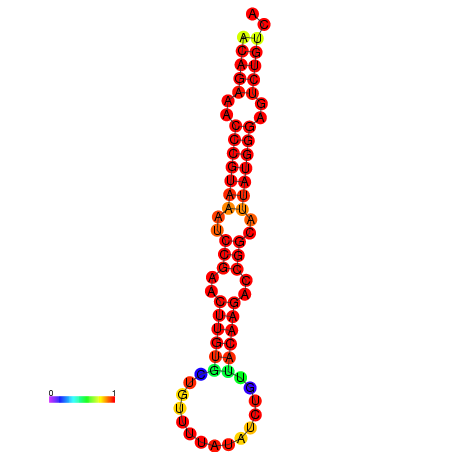 |
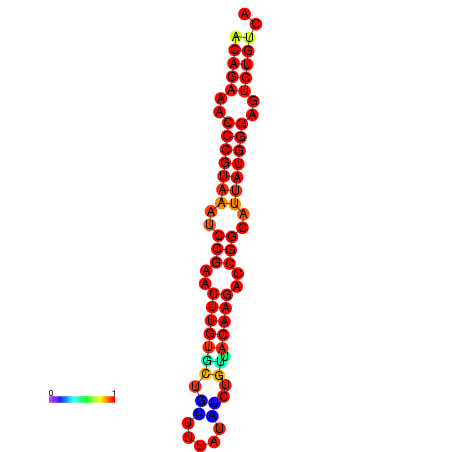 |
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
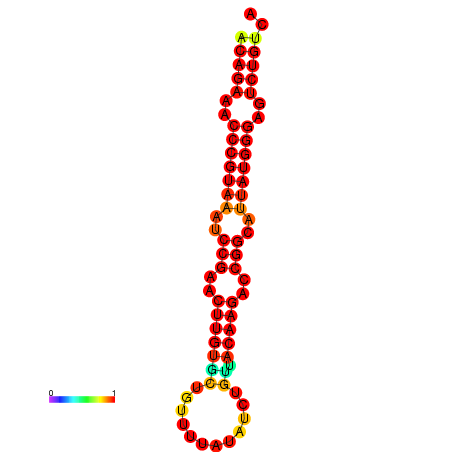 |
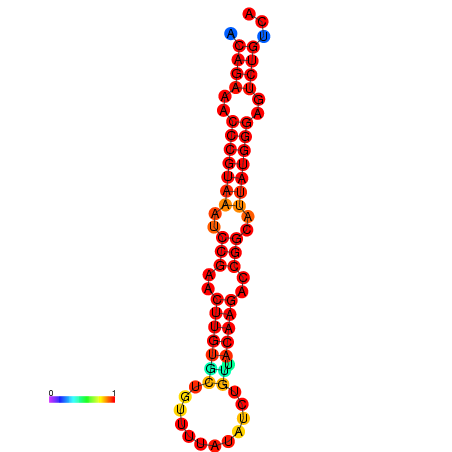 |
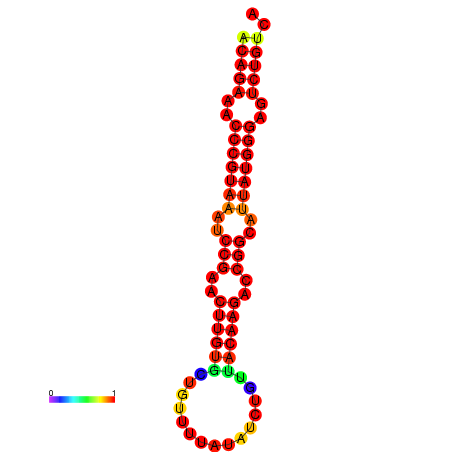 |
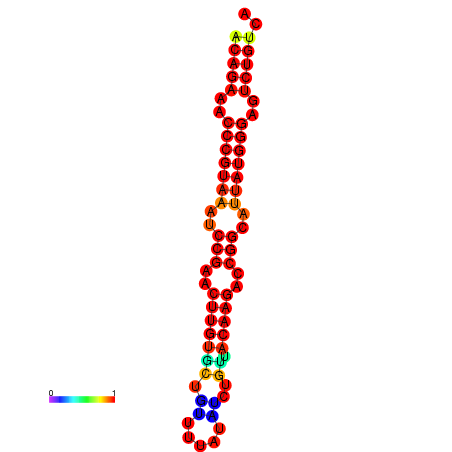 |
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
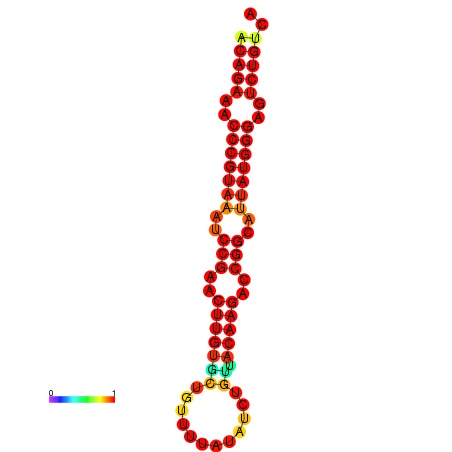 |
 |
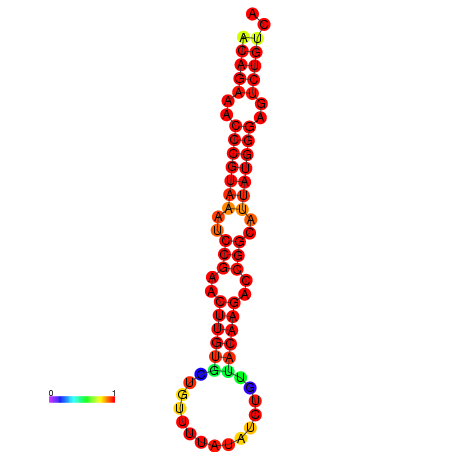 |
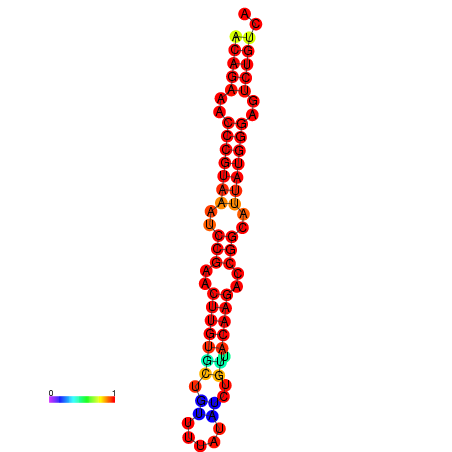 |
| dG=-23.8, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|
 |
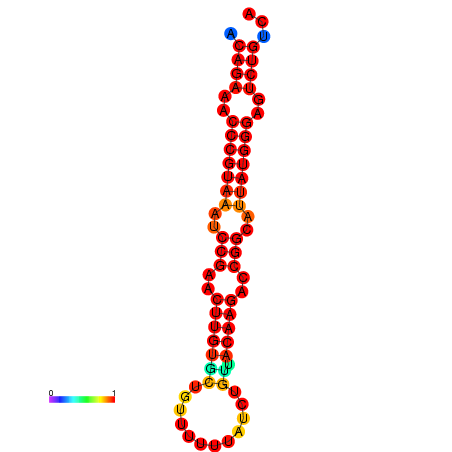 |
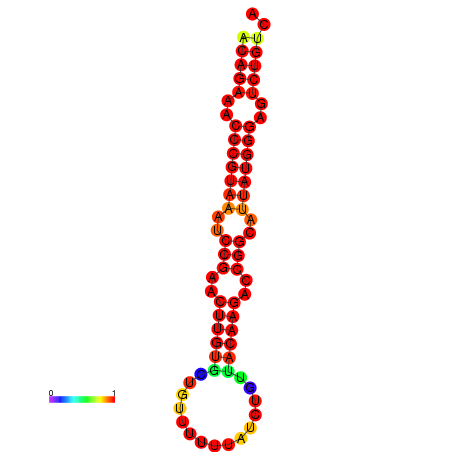 |
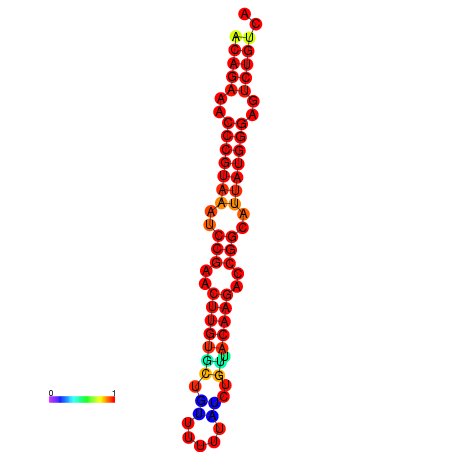 |
Generated: 03/07/2013 at 04:44 PM