| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:20487426-20487546 + | AAAGACC-----------------ATCCCAT--ATTCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ATTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G----AGTTGAAA |
| droSim1 | chr2L:20052694-20052814 + | AAAGACC-----------------ATCCCAT--CTTCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G----AGTTGAAA |
| droSec1 | super_18:365651-365771 + | AAGGACC-----------------ATCCCAT--CTTCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G----AGTTGAAA |
| droYak2 | chr2R:6842709-6842830 + | AAAGAAC-----------------ATCCCAC--CATCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGTCATCG----------------------------------G----AGTTGAAA |
| droEre2 | scaffold_4845:3954268-3954372 - | AAGGACC---------------------------TCCAGCCT-GTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G------------ |
| droAna3 | scaffold_12916:6985813-6985928 + | AAATATC-------------------------AAGAGAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAA-TATCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCAAGACTTCA----------------------------------A----GTTTAAAT |
| dp4 | chr4_group3:7845311-7845422 - | GGAAA--------------------------TCAGAAGGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAA-CATAAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCAAGACAGCG-----------------------------------------TCTACA |
| droPer1 | super_1:9292009-9292126 - | AAAGACGG-A-------------A-------TCAGAAGGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAA-CATAAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCAAGACATCG----------------------------------C------CTACAG |
| droWil1 | scaffold_180708:1845911-1846049 + | ATAGATGGTAGATATATATGTAG--------CTACTTGGCCTCTTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATATTAAATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGTCCGACATCA----------------------------------TCTACAATATAAT |
| droVir3 | scaffold_12963:13613413-13613527 - | TGTGC--------------------------TTACTTGGCCT-TTGAGAGTTCCATGCTTCCTTACATTCAATAGTATTC-AAATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAAACATCA----------------------------------C----AAAAACAA |
| droMoj3 | scaffold_6500:24555167-24555325 + | TGGGTCTT-AAA--------TTGT-------TCATTGCGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATTT-ATATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAAGCATCATACCAAACTGTAAAAAAAAACACATTATGAAATTG----TGTTTAAA |
| droGri2 | scaffold_15126:6250403-6250508 + | ATTGT--------------------------TTACTTGGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAT-AAATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAAACATC------------------------------------------------ |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:20487426-20487546 + | AAAGACC-----------------ATCCCAT--ATTCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ATTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G----AGTTGAAA |
| droSim1 | chr2L:20052694-20052814 + | AAAGACC-----------------ATCCCAT--CTTCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G----AGTTGAAA |
| droSec1 | super_18:365651-365771 + | AAGGACC-----------------ATCCCAT--CTTCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G----AGTTGAAA |
| droYak2 | chr2R:6842709-6842830 + | AAAGAAC-----------------ATCCCAC--CATCAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGTCATCG----------------------------------G----AGTTGAAA |
| droEre2 | scaffold_4845:3954268-3954372 - | AAGGACC---------------------------TCCAGCCT-GTGAGAGTTCCATGCTTCCTTGCATTCAATAGT-TAT-ACTCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAGACATCG----------------------------------G------------ |
| droAna3 | scaffold_12916:6985813-6985928 + | AAATATC-------------------------AAGAGAGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAA-TATCAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCAAGACTTCA----------------------------------A----GTTTAAAT |
| dp4 | chr4_group3:7845311-7845422 - | GGAAA--------------------------TCAGAAGGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAA-CATAAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCAAGACAGCG-----------------------------------------TCTACA |
| droPer1 | super_1:9292009-9292126 - | AAAGACGG-A-------------A-------TCAGAAGGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAA-CATAAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCAAGACATCG----------------------------------C------CTACAG |
| droWil1 | scaffold_180708:1845911-1846049 + | ATAGATGGTAGATATATATGTAG--------CTACTTGGCCTCTTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATATTAAATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGTCCGACATCA----------------------------------TCTACAATATAAT |
| droVir3 | scaffold_12963:13613413-13613527 - | TGTGC--------------------------TTACTTGGCCT-TTGAGAGTTCCATGCTTCCTTACATTCAATAGTATTC-AAATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAAACATCA----------------------------------C----AAAAACAA |
| droMoj3 | scaffold_6500:24555167-24555325 + | TGGGTCTT-AAA--------TTGT-------TCATTGCGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATTT-ATATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAAGCATCATACCAAACTGTAAAAAAAAACACATTATGAAATTG----TGTTTAAA |
| droGri2 | scaffold_15126:6250403-6250508 + | ATTGT--------------------------TTACTTGGCCT-TTGAGAGTTCCATGCTTCCTTGCATTCAATAGTATAT-AAATAAGCATATGGAATGTAAAGAAGTATGGAGCGAAATCTGGCGAAACATC------------------------------------------------ |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
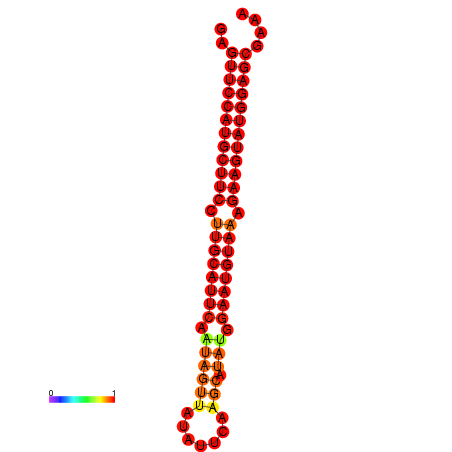
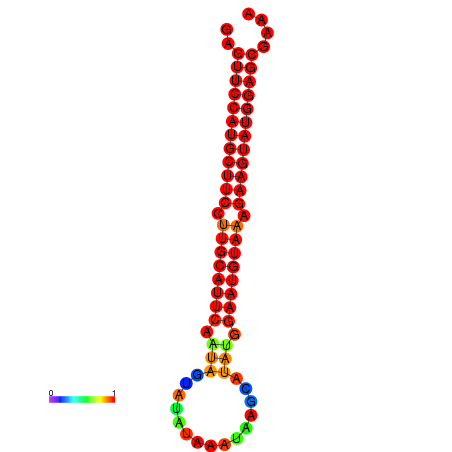
| dG=-29.0, p-value=0.009901 | dG=-28.3, p-value=0.009901 |
|---|---|
 |
 |
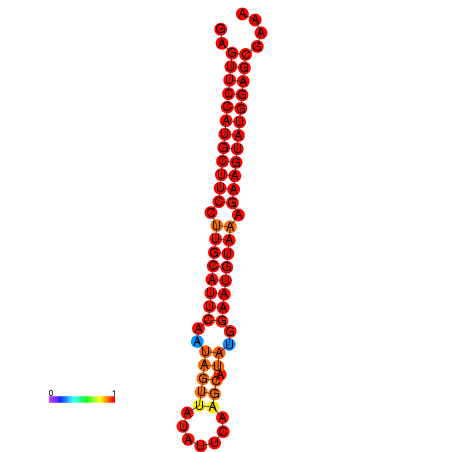
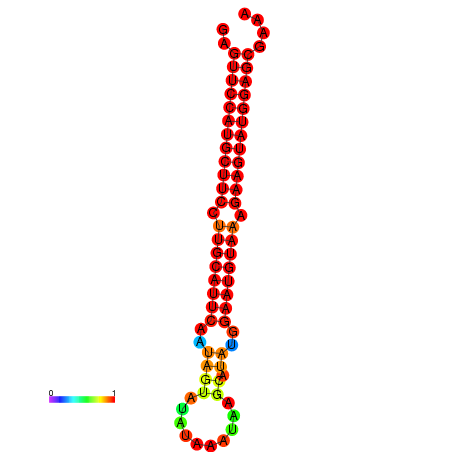
| dG=-29.0, p-value=0.009901 | dG=-28.3, p-value=0.009901 |
|---|---|
 |
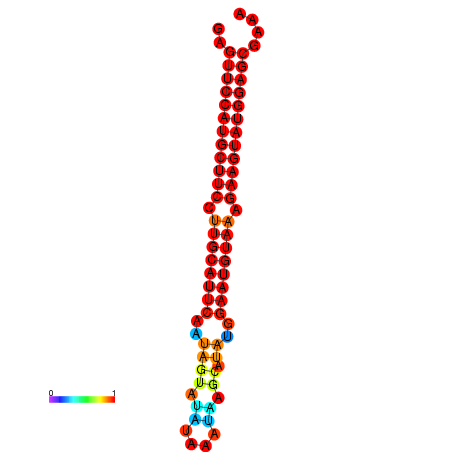
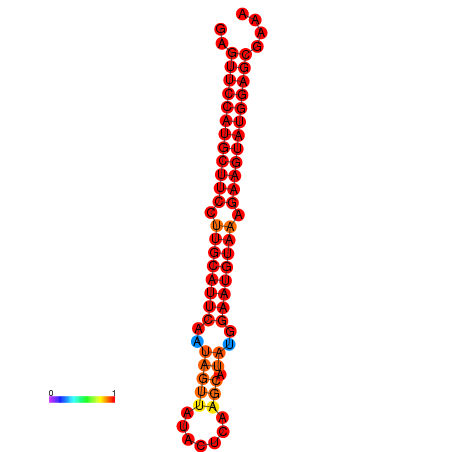 |
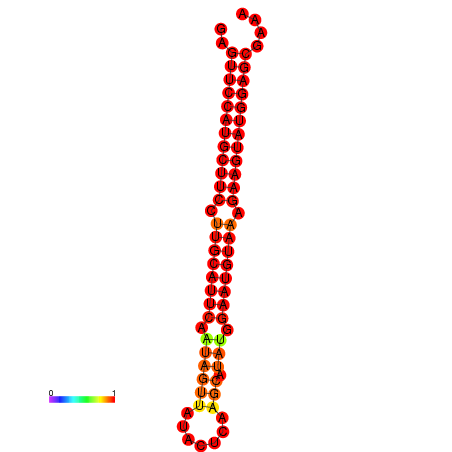
| dG=-29.0, p-value=0.009901 | dG=-28.3, p-value=0.009901 |
|---|---|
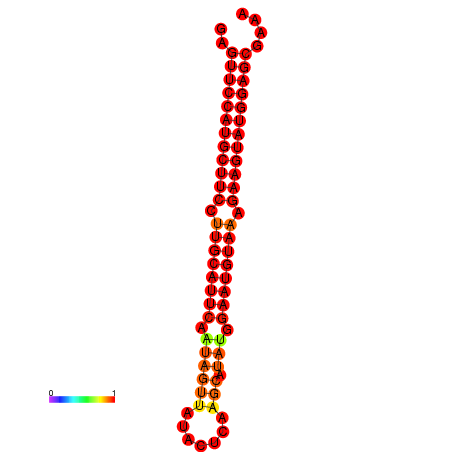 |
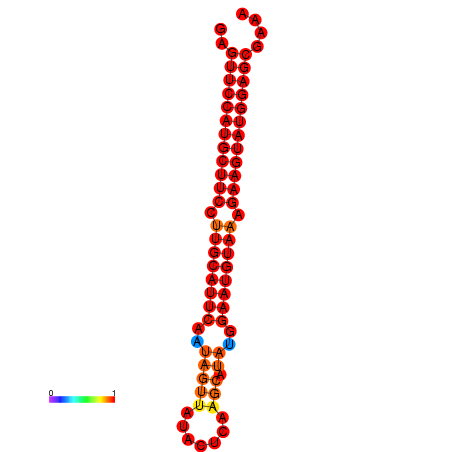 |
| dG=-27.9, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|---|
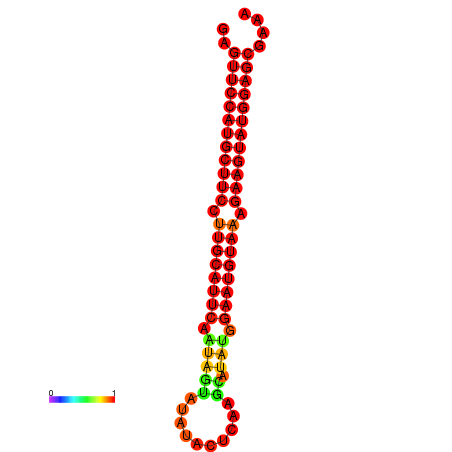 |
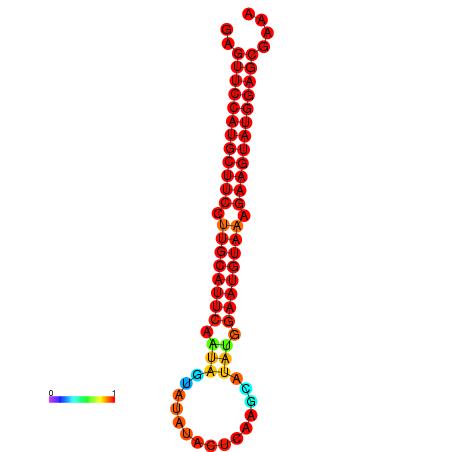 |
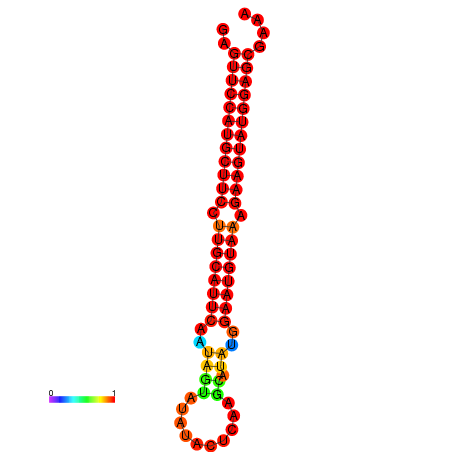 |
| dG=-29.0, p-value=0.009901 | dG=-28.3, p-value=0.009901 |
|---|---|
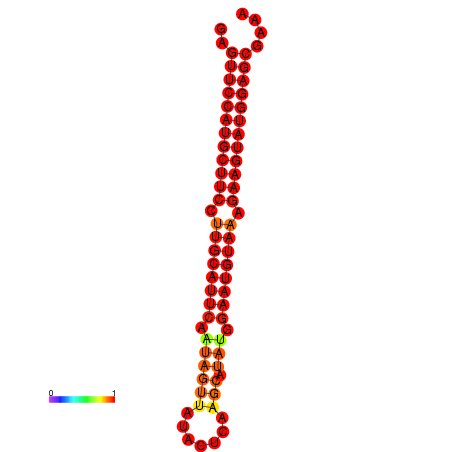 |
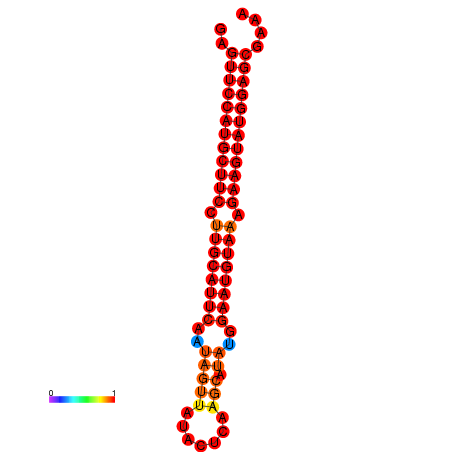 |
| dG=-27.9, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|---|
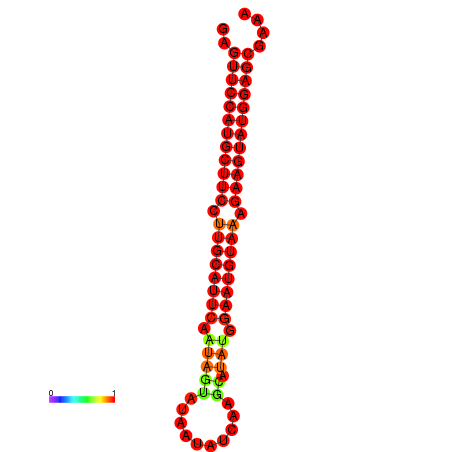 |
 |
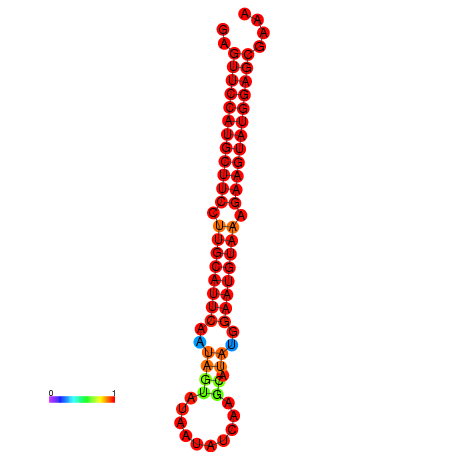 |
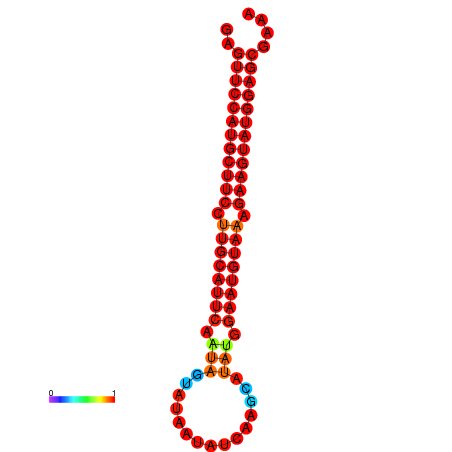
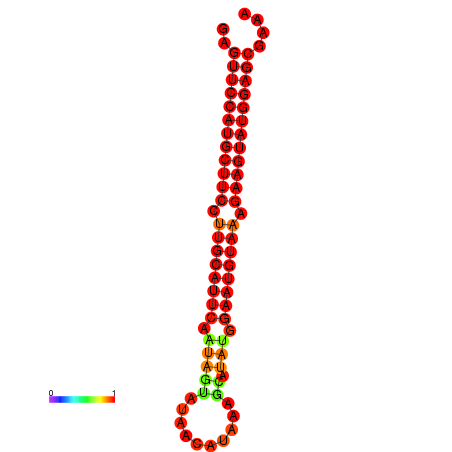
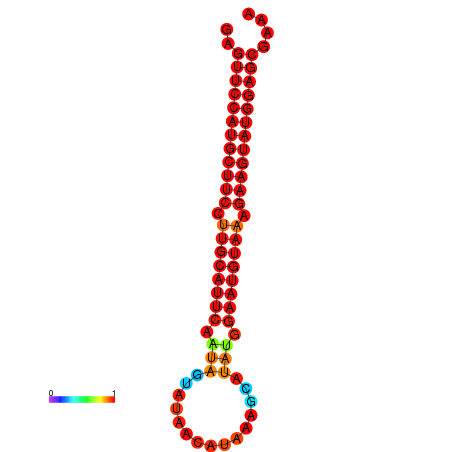
| dG=-27.9, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|---|
 |
 |
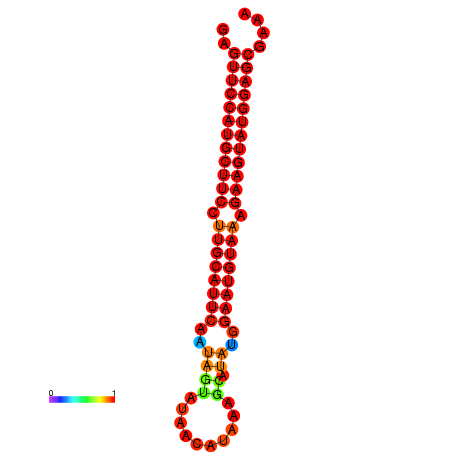 |
| dG=-27.9, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|---|
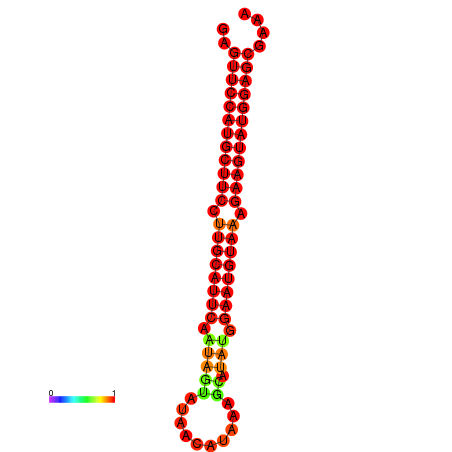 |
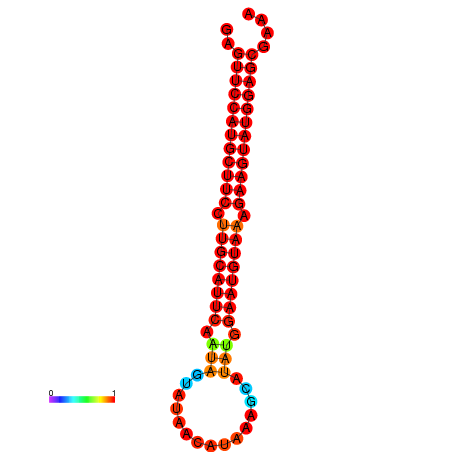 |
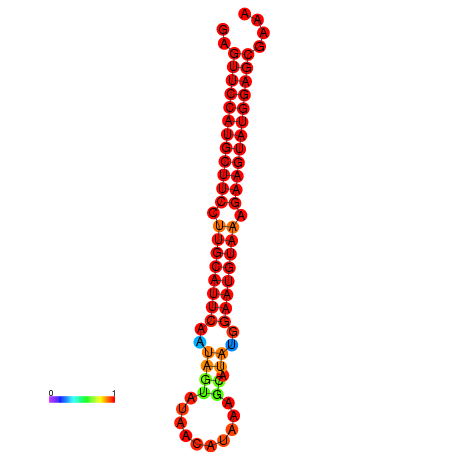 |
| dG=-28.5, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.7, p-value=0.009901 |
|---|---|---|---|
 |
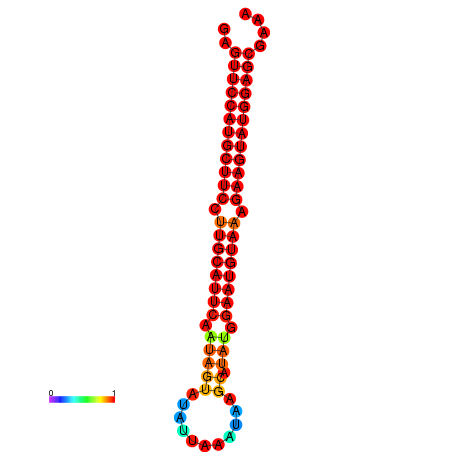 |
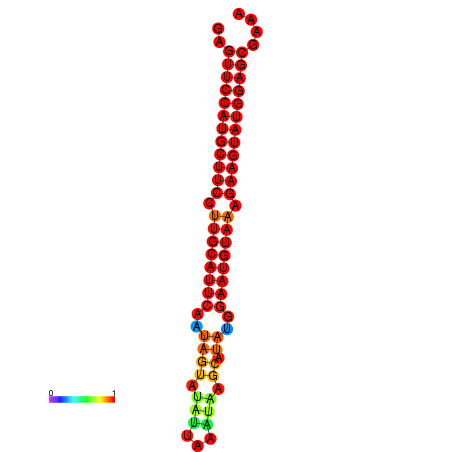 |
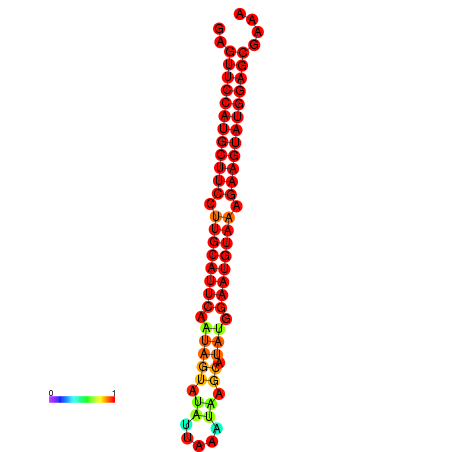 |
| dG=-27.9, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-26.9, p-value=0.009901 |
|---|---|---|---|
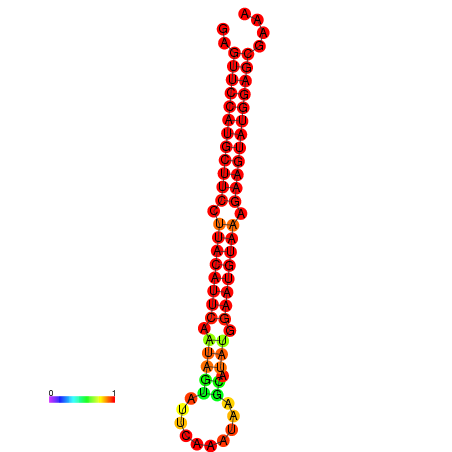 |
 |
 |
 |
| dG=-27.9, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-27.9, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:44 PM