dme-mir-995
No Changes
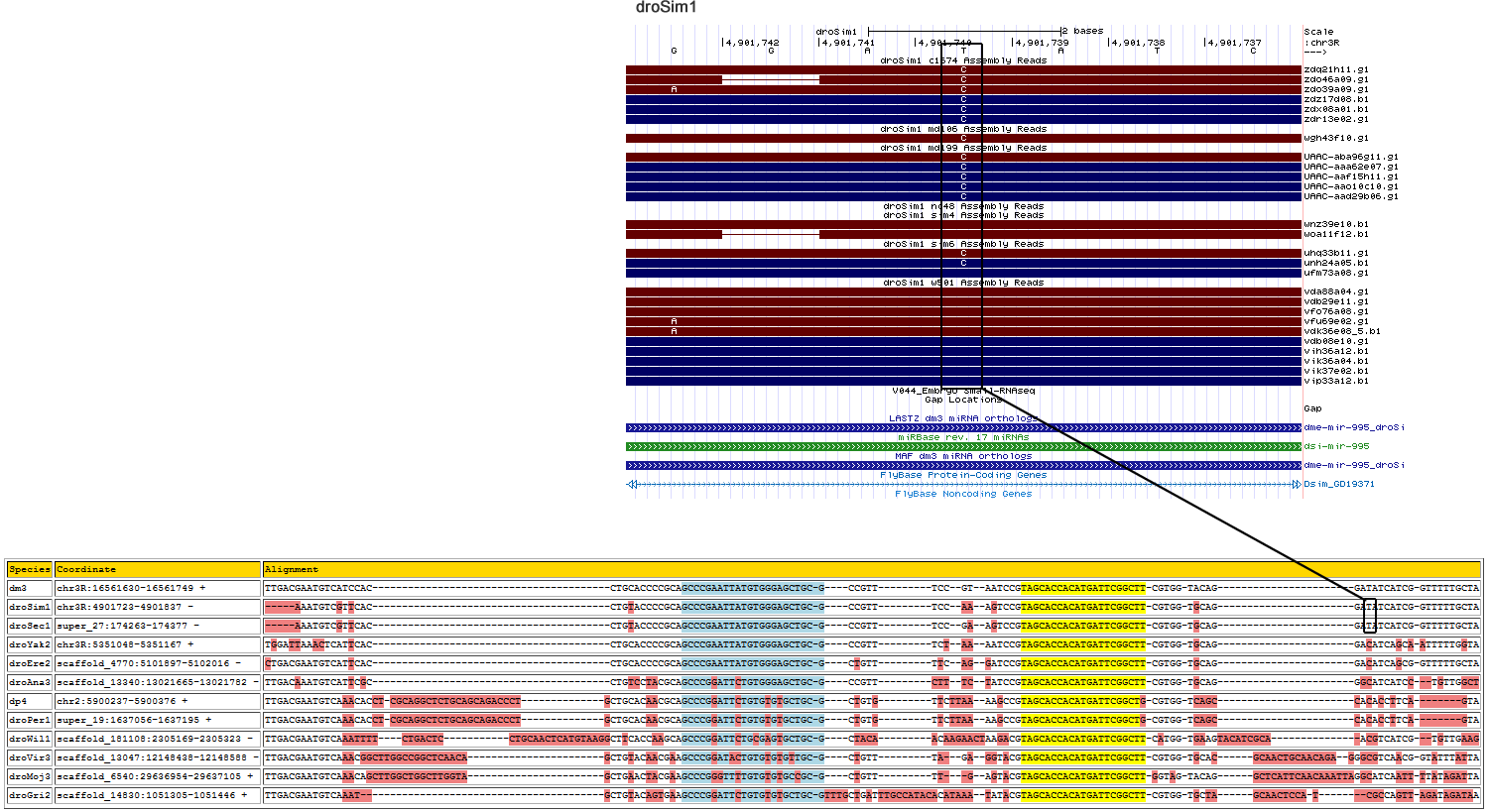
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr3R | 4901740 | A | 3 | 0 | 0 | 0 | 0 | 0 | 0.481 | 0 | 0.519 | 0 | 27 | Ambiguous - Suggested base change does not agree with neighboring species |
5' MOR |

Uncorrected Alignment:
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:16561630-16561749 + | TTGACG---------------AATGTCA-------------------------TCCAC--------------------------CTGCACCCCGCAGCCCGAATTATGTGGGAGCTGCG----CCGTTTCC-----------GTA----ATCCGTAGCACCACATGATTCGGCTTCGTGGTACA-----------------GGATATCATCGGT-TTTTGCTA |
| droSim1 | chr3R:4901723-4901838 - | -----A---------------AATGTCG-------------------------TTCAC--------------------------CTGTACCCCGCAGCCCGAATTATGTGGGAGCTGCG----CCGTTTCC-----------AAA----GTCCGTAGCACCACATGATTCGGCTTCGTGGTGCA-----------------GGATATCATCGGT-TTTTGCTA |
| droSec1 | super_27:174263-174377 - | ---------------------AATGTCG-------------------------TTCAC--------------------------CTGTACCCCGCAGCCCGAATTATGTGGGAGCTGCG----CCGTTTCC-----------GAA----GTCCGTAGCACCACATGATTCGGCTTCGTGGTGCA-----------------GGATATCATCGGT-TTTTGCTA |
| droYak2 | chr3R:5351033-5351167 + | TGGACGTTGCCATCCTGGATTAAACTCA-------------------------TTCAC--------------------------CTGCACCCCGCAGCCCGAATTATGTGGGAGCTGCG----CCGTTTCT-----------AAA----ATCCGTAGCACCACATGATTCGGCTTCGTGGTGCA-----------------GGACATCAGCAAT-TTTTGGTA |
| droEre2 | scaffold_4770:5101897-5102016 - | CTGACG---------------AATGTCA-------------------------TTCAC--------------------------CTGCACCCCGCAGCCCGAATTATGTGGGAGCTGCG----CTGTTTTC-----------AGG----ATCCGTAGCACCACATGATTCGGCTTCGTGGTGCA-----------------GGACATCAGCGGT-TTTTGCTA |
| droAna3 | scaffold_13340:13021664-13021782 - | TTGACA---------------AATGTCA-------------------------TTCGC--------------------------CTGTCCTACGCAGCCCGGATTCTGTGGGAGCTGCG----CCGTTCTT-----------TCT----ATCCGTAGCACCACATGATTCGGCTTCGTGGTGCA-----------------GGGCATCATCC-T-GTTGGCTG |
| dp4 | chr2:5900237-5900376 + | TTGACG---------------AATGTCA-------------------------AACACCTC-GCAGGCTCTGCAGCAGACCCTGCTGCACAACGCAGCCCGGATTCTGTGTGTGCTGCG----CTGTGTTC---------TTAAA----AGCCGTAGCACCACATGATTCGGCTGCGTGGTCAG-----------------CCACACCTTCAG--------TA |
| droPer1 | super_19:1637056-1637197 + | TTGACG---------------AATGTCA-------------------------AACACCTC-GCAGGCTCTGCAGCAGACCCTGCTGCACAACGCAGCCCGGATTCTGTGTGTGCTGCG----CTGTGTTC---------TTAAA----AGCCGTAGCACCACATGATTCGGCTGCGTGGTCAG-----------------CCACACCTTCAGT-ATC----- |
| droWil1 | scaffold_181108:2305175-2305323 - | TTGACG---------------AATGTCAAATTTTCTGACTCCTGCAACTCATGTAAGG--------------------------CTTCACCAAGCAGCCCGGATTCTGCGAGTGCTGCG----CTACAACA-----------AGAACTAAGACGTAGCACCACATGATTCGGCTTCATGGTGAA-----------------GTACATCGCAACG-TCATCGTG |
| droVir3 | scaffold_13047:12148438-12148588 - | TTGACG---------------AATGTCA-------------------------AACGGCTTGGCCGGCTCAACA---------GCTGTACAACGAAGCCCGGATACTGTGTGTGTTGCG----CTGTTTA------------GAG----GTACGTAGCACCACATGATTCGGCTTCGTGGTGCACGCAACTGCAACAGA--GGGCGTCAACGGT-ATTTATTA |
| droMoj3 | scaffold_6540:29636954-29637102 + | TTGACG---------------AATGTCA-------------------------AACAGCTTGGCTGGCTTGGTA---------GCTGAACTACGAAGCCCGGGTTTTGTGTGTGCCGCG----CTGTTTTG-------------A----GTACGTAGCACCACATGATTCGGCTTGGTAGTACAGGCTCATTCAACAAATTAGG---CATCAAT-TTTATAGA |
| droGri2 | scaffold_14830:1051305-1051441 + | TTGACG---------------AATGTCA-------------------------AAT---------------------------GCTGTACAGTGAAGCCCGGATTCTGTGTGTGCTGCGTTTGCTGATTTGCCATACACATAAAT----ATACGTAGCACCACATGATTCGGCTTCGTGGTGCTAG---------------CAACTCCATCGCCAGTTAGATA |