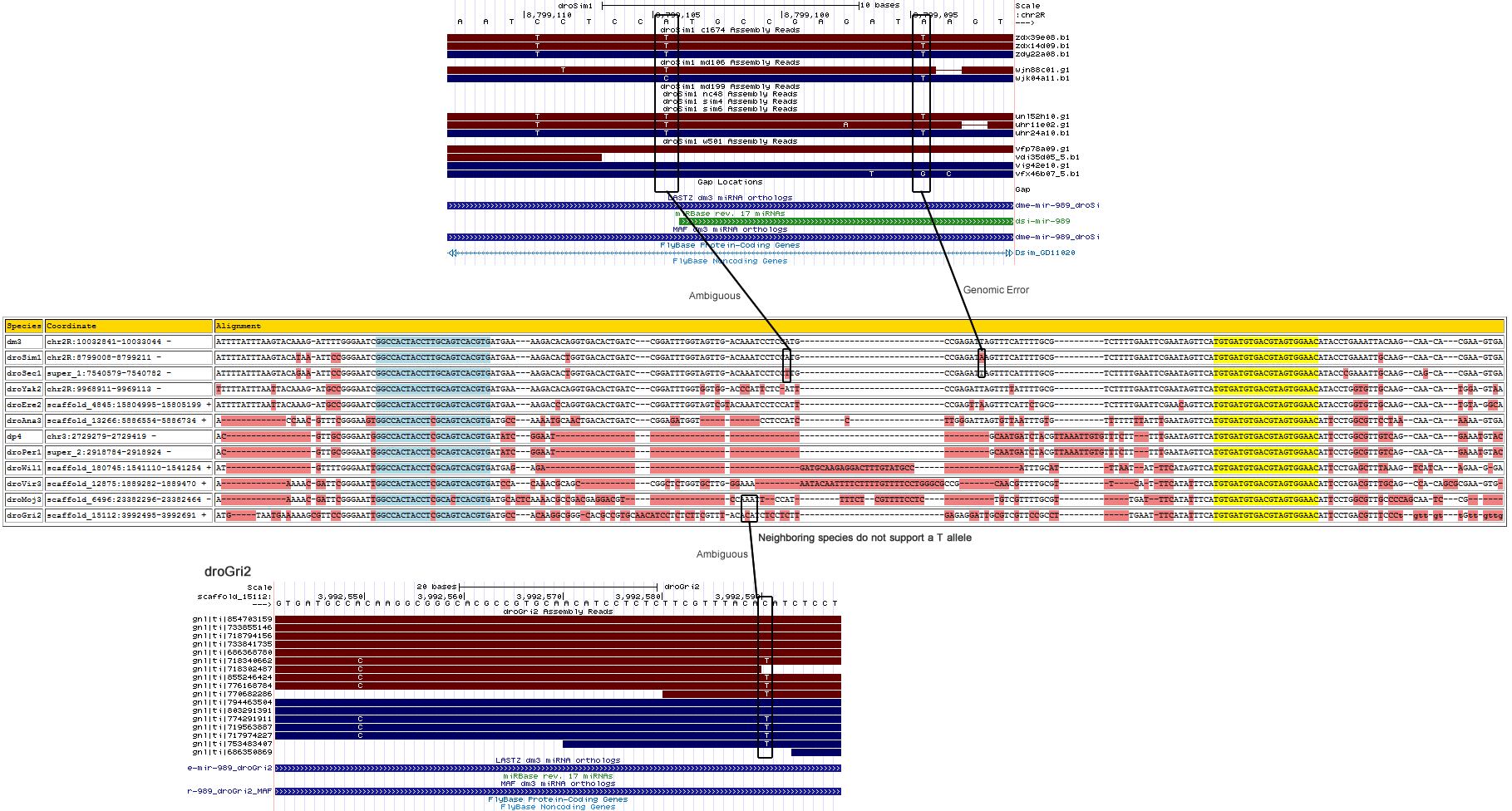
dme-mir-989
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droGri2 | scaffold_15112 | 3992591 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.467 | 0 | 0.533 | 15 | Ambiguous - Suggested base change does not agree with neighboring species |
Loop |
| droSim1 | chr2R | 8799095 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.545 | 0.091 | 0 | 0.364 | 11 | Genomic Error | Loop |
| droSim1 | chr2R | 8799105 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.636 | 0 | 0.091 | 0.273 | 11 | Ambiguous - The two alleles agree with either melanogaster or sechellia. |
Loop |

Corrected Alignment:
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:10032841-10033044 - | ATT-TTATTTAAGTACAA-------AGATTTTGGGAATCGGCCACTACCTTGCAGTCACGTGATGAA---AAGACACAGGTGACACTGATC---CGGATTTGGTAGTTG-ACAAATCCTCCATG-----------------------------CCGAGATTAGTTTCATTTTGCGTCTTTTGAA------TTCGAATAGTTCATGTGATGTGACGTAGTGGAACATACCTGAAATTACAAG-CAACA------CGAA-GT---GA |
| droSim1 | chr2R:8799008-8799211 - | ATT-TTATTTAAGTACAT-------AAATTCCGGGAATCGGCCACTACCTTGCAGTCACGTGATGAA---AAGACACTGGTGACACTGATC---CGGATTTGGTAGTTG-ACAAATCCTCCATG-----------------------------CCGAGATTAGTTTCATTTTGCGTCTTTTGAA------TTCGAATAGTTCATGTGATGTGACGTAGTGGAACATACCTGAAATTGCAAG-CAACA------CGAA-GT---GA |
| droSec1 | super_1:7540579-7540782 - | ATT-TTATTTAAGTACAG-------AAATTCCGGGAATCGGCCACTACCTTGCAGTCACGTGATGAA---AAGACACTGGTGACACTGATC---CGGATTTGGTAGTTG-ACAAATCCTCCTTG-----------------------------CCGAGATTAGTTTCATTTTGCGTCTTTTGAA------TTCGAATAGTTCATGTGATGTGACGTAGTGGAACATACCCGAAATTGCAAG-CAGCA------CGAA-GT---GA |
| droYak2 | chr2R:9968911-9969114 - | TTTTTTATTTAATTACAA-------AGATGCCGGGAATCGGCCACTACCTTGCAGTCACGTGATGAA---AAGACACAGGTGACACTGATC---CGGATTTGGTGGTGG-ACCCATTCTC-ATT-----------------------------CCGAGATTAGTTTTATTTTGCGTCTTTTGAA------TTCGAATAGTTCATGTGATGTGACGTAGTGGAACATACCTGGTGTTGCAAG-CAACA------TGGA-GT---AA |
| droEre2 | scaffold_4845:15804995-15805201 + | ATT-TTATTTAATTACAA-------AGATGCCGGGAATCGGCCACTACCTTGCAGTCACGTGATGAA---AAGACCCAGGTGACACTGATC---CGGATTTGGTAGTCGTACAAATCCTCCATT-----------------------------CCGAGTTAAGTTTCATTCTGCGTCTTTTGAA------TTCGAACAGTTCATGTGATGTGACGTAGTGGAACATACCTGGTGTTGCAAG-CAACA------TGTA-GGCA-TA |
| droAna3 | scaffold_13266:5886554-5886734 + | A--------------CCA-------ACGTTTCGGGAAGTGGCCACTACCTCGCAGTCACGTGATGCC---AAAATGCAACTGACACTGATC---CGGAGATGGT------------CCTCCATC---------C-------------------TTGGGATTAGTGTTAATTTGTGTTTTTTTTA------TTTGAATAGTTCATGTGATGTGACGTAGTGGAACATTCCTGGCGTTCCTAA-CAACA------AAAA-GT---GA |
| dp4 | chr3:2729279-2729420 - | A-------------------------CGTTGCGGGAATGGGCCACTACCTCGCAGTCACGTGATATC---GGAATGCAA------------------------------------T-----------------------------------------GATCTACGTTAAATTGTGTTTCTT---------TTTGAATAGTTCATGTGATGTGACGTAGTGGAACATTCCTGGCGTTGTCAG-CAACA------GAAATGT---AC |
| droPer1 | super_2:2918784-2918948 - | TTG-TAAACCAAGAAGAACCAAACAACGTTGCGGGAATGGGCCACTACCTCGCAGTCACGTGATATC---GGAATGCAA------------------------------------T-----------------------------------------GATCTACGTTAAATTGTGTTTCTT---------TTTGAATAGTTCATGTGATGTGACGTAGTGGAACATTCCTGGCGTTGTCAG-CAACA------GAAATGT---AC |
| droWil1 | scaffold_180745:1541090-1541255 + | TAC-GAGTTCGAGTACAA----TAAATGTTTTGGGAATTGGCCACTACCTCGCAGTCACGTGATGAG---AGA---------------------------------------------------GATGCAAGA------------------------GGACTTTG------TATGCCATTTGCATTTAATATTTCATAGTTCATGTGATGTGACGTAGTGGAACATTCCTGAGCTTTAAAGTCATCA------AGAA-GG---AA |
| droVir3 | scaffold_12875:1889282-1889470 + | A---------------------AAAACGATTCGGGAATTGGCCACTACCTCGCAGTCACGTGATCCA---CAAACGCAGC--------------CGGCTCTGGTGCTTG-GGAAA---------AATACAATTTTCTTTTGTTTTCCTGGGCGCCGCA-------ACGTTTTGCGT----TCA--------TTTCATATTTCATGTGATGTGACGTAGTGGAACATTCCTGACGTTTGCAG-CCACA------GCGC-GAAGTG- |
| droMoj3 | scaffold_6496:23382296-23382464 - | A---------------------AAAACGATTCGGGAATTGGCCACTACCTCGCACTCACGTGATGCACTCAAAACGCCGACGAGGACGT---------------------CCAAATT--CCAT---------TTTCT--CGTTTTCCTC--------------TGTCGTTTTGCGT----TGA--------TTTCATATTTCATGTGATGTGACGTAGTGGAACATTCCTGGCGTTGCC-----CCAGCAATCCG---------- |
| droGri2 | scaffold_15112:3992495-3992691 + | ATG-TAAT-----GAA------AAAGCGTTCCGGGAATTGGCCACTACCTCGCAGTCACGTGATGCC---ACAAGGCGGGC-ACGCCGTGCAACATCCTCTCTTCGTTT-ACACATCTCCTCTT-----------------------------GAGAGGATTGCGTCGTTCCGCCT----TGAA-------TTTCATATTTCATGTGATGTGACGTAGTGGAACATTCCTGACGTTTCCCA-CAACA------AGAA-CA---AC |