dme-mir-986
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
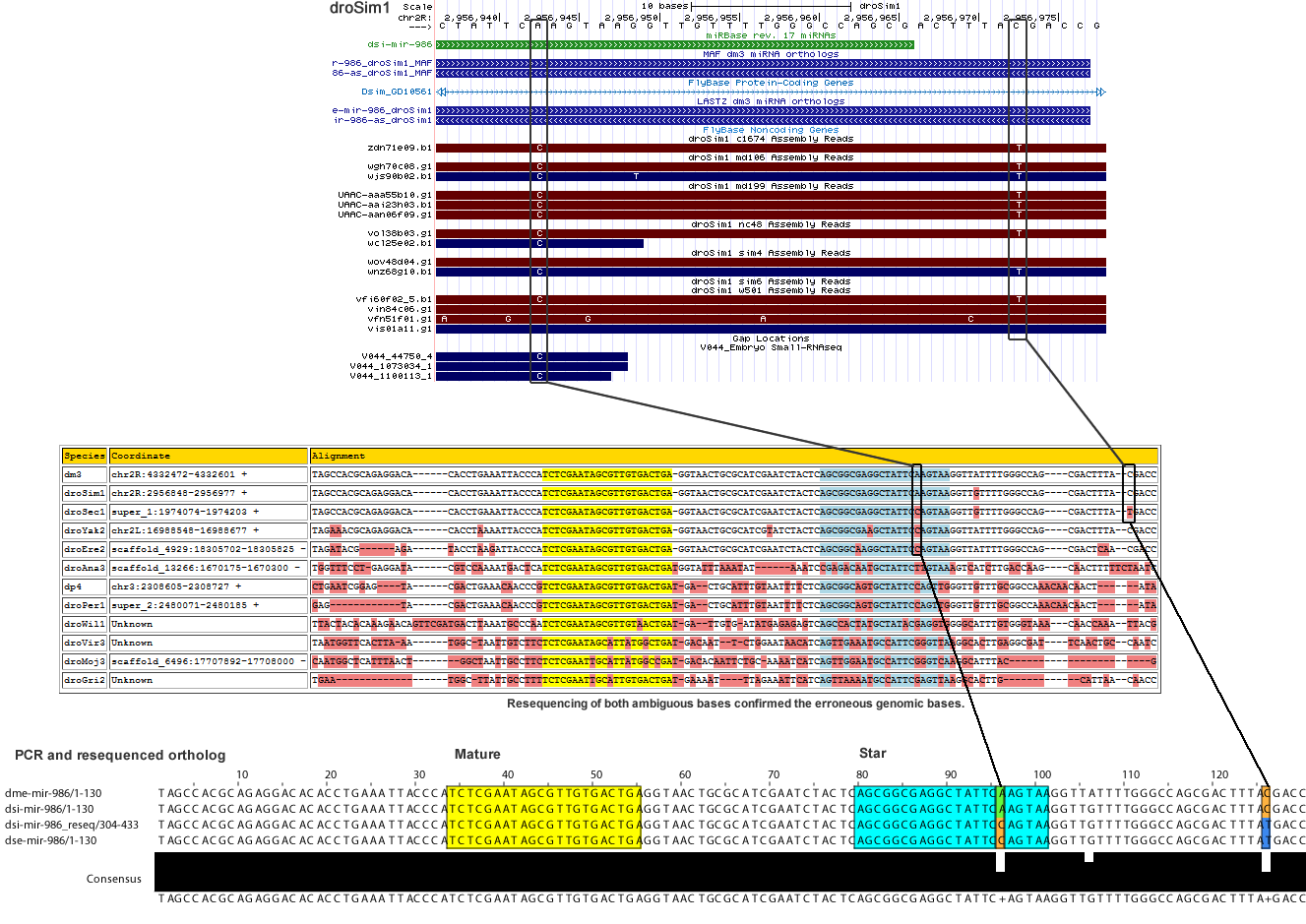
| droSim1 | chr2R | 2956943 | A | 3 | 0.167 | 0.833 | 0 | 0 | 6 | 0.286 | 0.714 | 0 | 0 | 14 | Ambiguous - substitution agrees with the sechellia ortholog rather than the simulans ortholog after resequencing. | mir* |
| droSim1 | chr2R | 2956973 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.308 | 0 | 0.692 | 13 | Ambiguous - substitution agrees with the sechellia ortholog rather than the simulans ortholog after resequencing. | 3' MOR |

Corrected Alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:4332472-4332601 + | TAGCCA-------CGCAG----AGGA-------CACAC-----CTGAA--------ATTACCCATCTCGAATAGCGTTGTGACTGAG-GTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCAAGTAAGGTTATTTTGGGCC----AGCGACTTT--ACGACC |
| droSim1 | chr2R:2956848-2956977 + | TAGCCA-------CGCAG----AGGA-------CACAC-----CTGAA--------ATTACCCATCTCGAATAGCGTTGTGACTGAG-GTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCCAGTAAGGTTGTTTTGGGCC----AGCGACTTT--ATGACC |
| droSec1 | super_1:1974074-1974203 + | TAGCCA-------CGCAG----AGGA-------CACAC-----CTGAA--------ATTACCCATCTCGAATAGCGTTGTGACTGAG-GTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCCAGTAAGGTTGTTTTGGGCC----AGCGACTTT--ATGACC |
| droYak2 | chr2L:16988548-16988677 + | TAGAAA-------CGCAG----AGGA-------CACAC-----CTAAA--------ATTACCCATCTCGAATAGCGTTGTGACTGAG-GTAACTGCGCATCGTATCTACTCAGCGGCGAAGCTATTCCAGTAAGGTTATTTTGGGCC----AGCGACTTT--ACGACC |
| droEre2 | scaffold_4929:18305702-18305825 - | TAGATA-------C----------GA-------GATAC-----CTAAG--------ATTACCCATCTCGAATAGCGTTGTGACTGAG-GTAACTGCGCATCGAATCTACTCAGCGGCAAGGCTATTCCAGTAAGGTTATTTTGGGCC----AGCGACTCA--ACGACC |
| droAna3 | scaffold_13266:1670175-1670300 - | TGGTTT--------CCTG----AGGA-------TACGT-----CCAAA--------ATGACTCATCTCGAATAGCGTTGTGACTGATGGTATTT------AAATATAAATCCGAGACAATGCTATTCTTGTAAAGTCATCTTGACCA----AGCAACTTTTTCTAATC |
| dp4 | chr3:2308599-2308727 + | TATCTC-------CT--GAATCGGAG-------TACGA-----CTGAA--------ACAACCCGTCTCGAATAGCGTTGTGACTGAT---GACTGCATTTGTAATTTTCTCAGCGGCAGTGCTATTCCAGTTGGGTTGTTTGCGGCCAAACAACAACTAT-------A |
| droPer1 | super_2:2480065-2480185 + | GAATC-----------------GGAG-------TACGA-----CTGAA--------ACAACCCGTCTCGAATAGCGTTGTGACTGAT---GACTGCATTTGTAATTTTCTCAGCGGCAGTGCTATTCCAGTTGGGTTGTTTGCGGCCAAACAACAACTAT-------A |
| droWil1 | scaffold_180700:3161017-3161152 - | TTACTACA-----CAAAG----TGAACAGTTC-GATGA-----CTTAA--------ATGCCCAATCTCGAATAGCGTTGTAACTGAT-G--ATTGTG-ATATGAGAGAGTCAGCCACTATGCTATACGAGGTGGGGCATTTGTGGGTA---AACAACCAA--ATTACG |
| droVir3 | scaffold_12875:9403376-9403510 + | AAACTATAAAGTGTTTTA----ATGG-------TTCAC-----TT-AAATGGCTAATTGTCTTCTCTCGAATAGCATTATGGCTGAT-GACAATT---CTGGAATAACATCAGTTGAAATGCCATTCGGGTTAAGGCACTTGAGAAATGGCAGCGA------------ |
| droMoj3 | scaffold_6496:17707880-17708014 - | TGTTGG-------CGAA-----CTTT-CAATGGCTCAT-----TTAA-CTGGCTAATTGCCTTCTCTCGAATTGCATTATGGCCGAT-GACACAATTCTGCAAAATC-ATCAGTTGGAATGCCATTCGGGTCAAGGCATTTACGAATCGCCAACGA------------ |
| droGri2 | scaffold_15245:2671305-2671410 + | TTGAAT-------GGC-----------------------------TTA--------TTGCCTTTTCTCGAATTGCATTGTGACTGAT-GAAAAT----TTAGAAATTCATCAGTTAAAATGCCATTCGAGTTAAGGCACTTATG-----------CATTA--ACAACC |