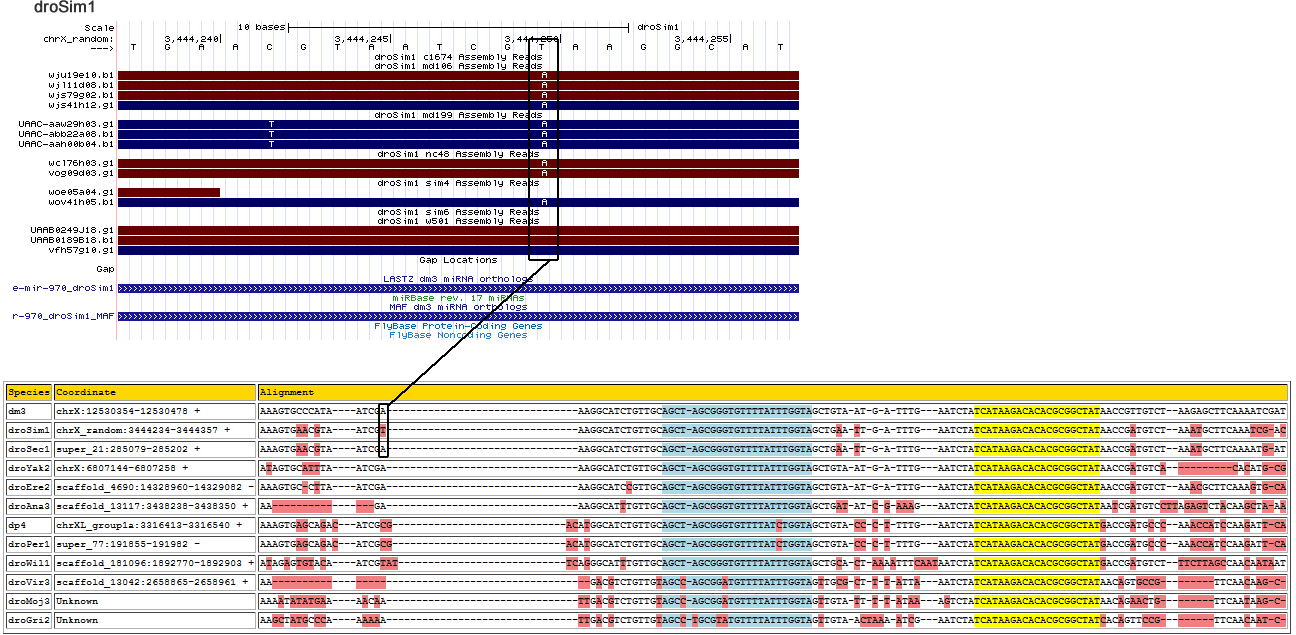
dme-mir-970
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chrX_random | 3444250 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.769 | 0 | 0 | 0.231 | 13 | Genomic Error | 5' MOR |

Corrected Alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:12530354-12530478 + | AAAGTGCCCAT-------------------AA------TCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGTAATG--A-TTTG---AATCTATCATAAGACACACGCGGCTATAACCGTT--GTCT--AAGAGCTTCAAA---A-------------------TC------GAT |
| droSim1 | chrX_random:3444234-3444357 + | AAAGTGAACGT-------------------AA------TCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGAATTG--A-TTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCT--AAATGCTTCAAA-----------------------TC------GAC |
| droSec1 | super_21:285079-285202 + | AAAGTGAACGT-------------------AA------TCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGAATTG--A-TTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCT--AAATGCTTCAAA---A--------------------T------GAT |
| droYak2 | chrX:6807144-6807273 + | ATAGTGCATTT-------------------AA------TCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGTAATG--A-TTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCA--C---------ACATGCGACACA--------------CACATTAGAT |
| droEre2 | scaffold_4690:14328958-14329082 - | AAAGTGC-CTT-------------------AA------TCGAAAGGCATCCGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGTAATG--A-TTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCT--AAACGCTTCAAA---G----TG--------------C------AAT |
| droAna3 | scaffold_13117:3438231-3438350 + | TTAGTGGA-------------------------------AGAAAGGCATTTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGATATC--G-AAAG---AATCTATCATAAGACACACGCGGCTATAATCGAT--GTCCTTAGAGTCTACAAG---C--------------------T------AAA |
| dp4 | chrXL_group1a:3316403-3316556 + | ATCACCC---CAAAAAGTGAGCAGACA-----TCGCG-A--CATGGCATCTGTTGCAGCTAGCGGGTGTTTTATCTGGTAGCTGTACCC--T-TTTG---AATCTATCATAAGACACACGCGGCTATGACCGAT--GCCC--AAACCATCCAAG---A----TTCAATAATTAGTAATC------AAT |
| droPer1 | super_77:191839-191992 - | ATCACCC---CAAAAAGTGAGCAGACA-----TCGCG-A--CATGGCATCTGTTGCAGCTAGCGGGTGTTTTATCTGGTAGCTGTACCC--T-TTTG---AATCTATCATAAGACACACGCGGCTATGACCGAT--GCCC--AAACCATCCAAG---A----TTCAATAATTAGTAATC------AAT |
| droWil1 | scaffold_181096:1892772-1892900 + | AGAGTGTAC---------------------AATCGTATT--CAGGGCATTTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGCACTA-AAATTTCAATAATCTATCATAAGACACACGCGGCTATGACCGAT--GTCTTT-----CT----------------------TAGCCAAC------AAT |
| droVir3 | scaffold_13042:2658865-2658967 + | --------------------------------------------GACGTCTGTTGTAGCCAGCGGATGTTTTATTTGGTAGTTGCGCTT--T-ATTA---AATCTATCATAAGACACACGCGGCTATAACAGTGCCG--TTCA---------ACAAGCAA------------------C------GTC |
| droMoj3 | scaffold_6328:939989-940116 - | TAAACCC---CA----GTGATCGACTATGAAA------ACAATTGACGTCTGTTGTAGCCAGCGGATGTTTTATTTGGTAGTTGTATTT--T-ATAA---AGTCTATCATAAGACACACGCGGCTATAACAGAACTG--TTCA---------ATA------------------------------AGC |
| droGri2 | scaffold_15203:5475247-5475378 + | AAAGTCTGCCC-------------------AA------AAAATTGACGTCTGTTGTAGCCTGCGTATGTTTTATTTGGTAGTTGTAACTAAA-ATCG---AATCTATCATAAGACACACGCGGCTATCACAGTTCCG--TTCA---------ACAATCAACGTA--------------C-CATT-ACA |