dme-mir-968
Changes
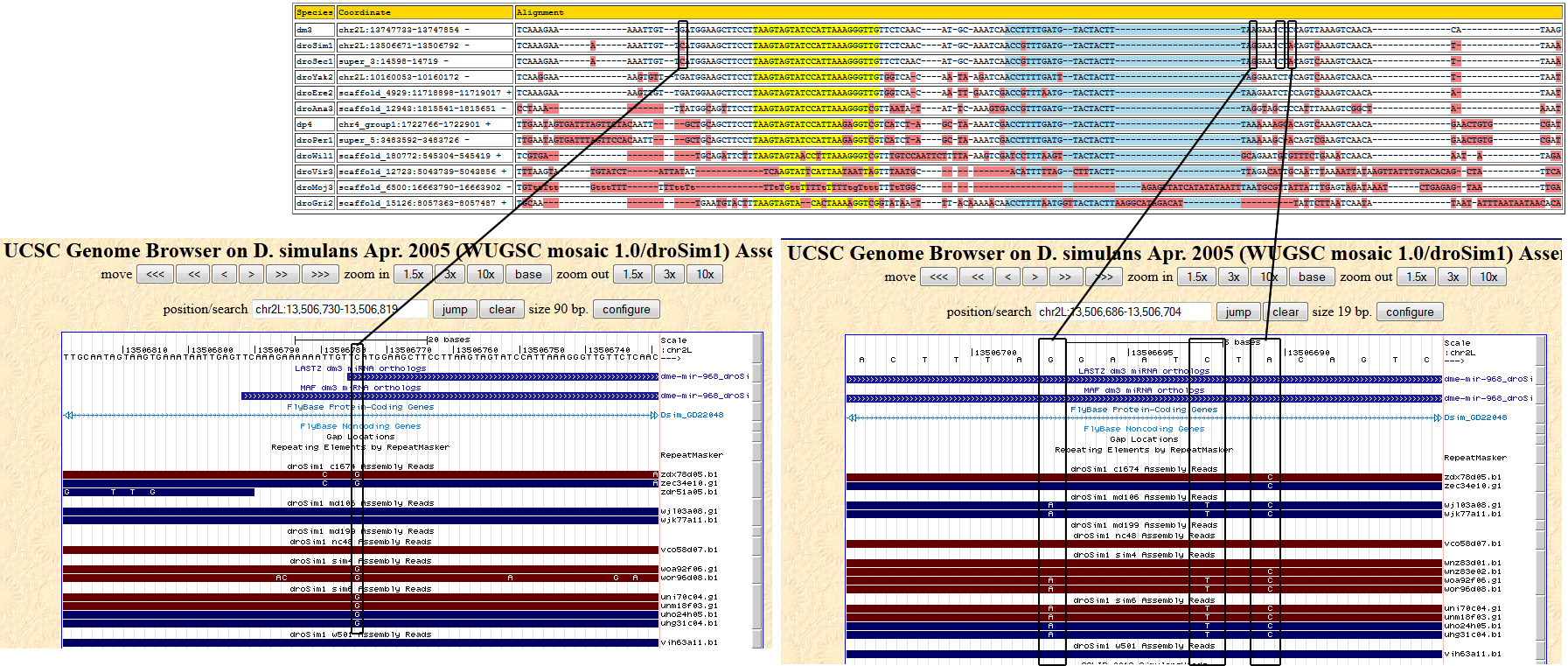
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr2L | 13506691 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.786 | 0.214 | 14 | Ambiguous - The two alleles agree with either melanogaster or sechellia. |
5' flanking region |
| droSim1 | chr2L | 13506693 | G | 3 | 0 | 0 | 0 | 0 | 0 | 0.571 | 0 | 0.429 | 0 | 14 | Ambiguous - Suggested base change does not agree with neighboring species |
5' flanking region |
| droSim1 | chr2L | 13506698 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.429 | 0 | 0.571 | 14 | Ambiguous - The two alleles agree with either melanogaster or sechellia. |
5' flanking region |
| droSim1 | chr2L | 13506775 | G | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0.667 | 0.333 | 0 | 12 | Ambiguous - The two alleles agree with either melanogaster or sechellia. |
3' flanking region |

Corrected
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:13747733-13747854 - | TCAAAGA-AA------------AATTGTTGATGGAAGCTTCCTTAAGTAGTATCCATTAAAGGGTTGTTCTCAACATG-----CAAATCAACCTTTTGATGTACTACTTTAAGAATCTCCAGTTAAAGTCAACACA--TAAG |
| droSim1 | chr2L:13506671-13506792 - | TCAAAGAAAA------------AATTGTTCATGGAAGCTTCCTTAAGTAGTATCCATTAAAGGGTTGTTCTCAACATG-----CAAATCAACCGTTTGATGTACTACTTTAGGAATCTACAGTCAAAGTCAACAT---TAAA |
| droSec1 | super_3:14598-14719 - | TCAAAGAAAA------------AATTGTTCATGGAAGCTTCCTTAAGTAGTATCCATTAAAGGGTTGTTCTCAACATG-----CAAATCAACCGTTTGATGTACTACTTTAGGAATCTACAGTCAAAGTCAACAT---TAAA |
| droYak2 | chr2L:10160053-10160172 - | TCAAGGA-AA------------AGTGTTTGATGGAAGCTTCCTTAAGTAGTATCCATTAAAGGGTTGTGGTCA-CAAT-----AAGATCAACCTTTTGATTTACTACTTTAGGAATCTCCAGTCAAAGTCAACAT---TAAT |
| droEre2 | scaffold_4929:11718898-11719017 + | TCAAAGA-AA------------AGTTGTTGATGGAAGCTTCCTTAAGTAGTATCCATTAAAGGGTTGTGGTCA-CAAT-----TGAATCGACCGTTTAATGTACTACTTTAAGAATCTCCAGTCAAAGTCAACAA---TAAT |
| droAna3 | scaffold_12943:1815538-1815649 - | AAA---------------------T----TATGGCAGTTTCCTTAAGTAGTATCCATTAAAGGGTCGTTAATA-TATT-----CAAAGTGACCGTTTGATGTACTACTTTAGGTAGCTCCATTTAAAGTCGGCTAAAATAAG |
| dp4 | chr4_group1:1722766-1722895 + | TTGAATAGTGATTTAGTTCTACAATT----GCTGCAGCTTCCTTAAGTAGTATCCATTAAGAGGTCGTCATCT-AGCT-----AAAATCGACCTTTTGATGTACTACTTTAAAAAAGCACAGTCAAAGTCAACAGAA--CTG |
| droPer1 | super_5:3483598-3483726 - | TTGAATAGTGATTTAGTTCCACAATT----GCTGCAGCTTCCTTAAGTAGTATCCATTAAGAGGTCGTCATCT-AGCT-----AAAATCGACCTTTTGATGTACTACTTTAAAAAGC-ACAGTCGAAGTCAACAGAA--CTG |
| droWil1 | scaffold_180772:545302-545417 + | CTCGTG------------------------A-TGCAGATTCTTTAAGTAGTAACCTTTAAAGGGTCGTTTGTCCAATTCTTTTAAAGTCGATCCTTTAAGTTACTACTTGCAGAATGTGTTTCTGAAATCAACAAA--TATA |
| droVir3 | scaffold_12723:5044036-5044131 - | C---------------------AATT----A-TACATGTGCCTTAAGTAGTAACCATTAAGAGGTTGTTGTTA-GATT-----TAAATCCACCTTTTAATGTACTACTTATGACAAATGCATAG--------------CAAA |
| droMoj3 | scaffold_6500:16664499-16664593 + | AAA--------------------ATT----A-TGCAAGTATCATAAGTAGTAACCATTAAAAGGTCGTGATT-----T-----GAACTCAACCTTTTAATTTACTACTTGTGATAAATGCATAGAAATGT------------ |
| droGri2 | scaffold_15126:8057347-8057450 - | TCTA----------------------------------TGCCTTAAGTAGTAACCATTAAAAGGTTGTTTTTGTAAAT-----TATACCGACCTTTTAGTGTACTACTTAAAGTACATTCATTGCAAGTCGACGAATATATA |