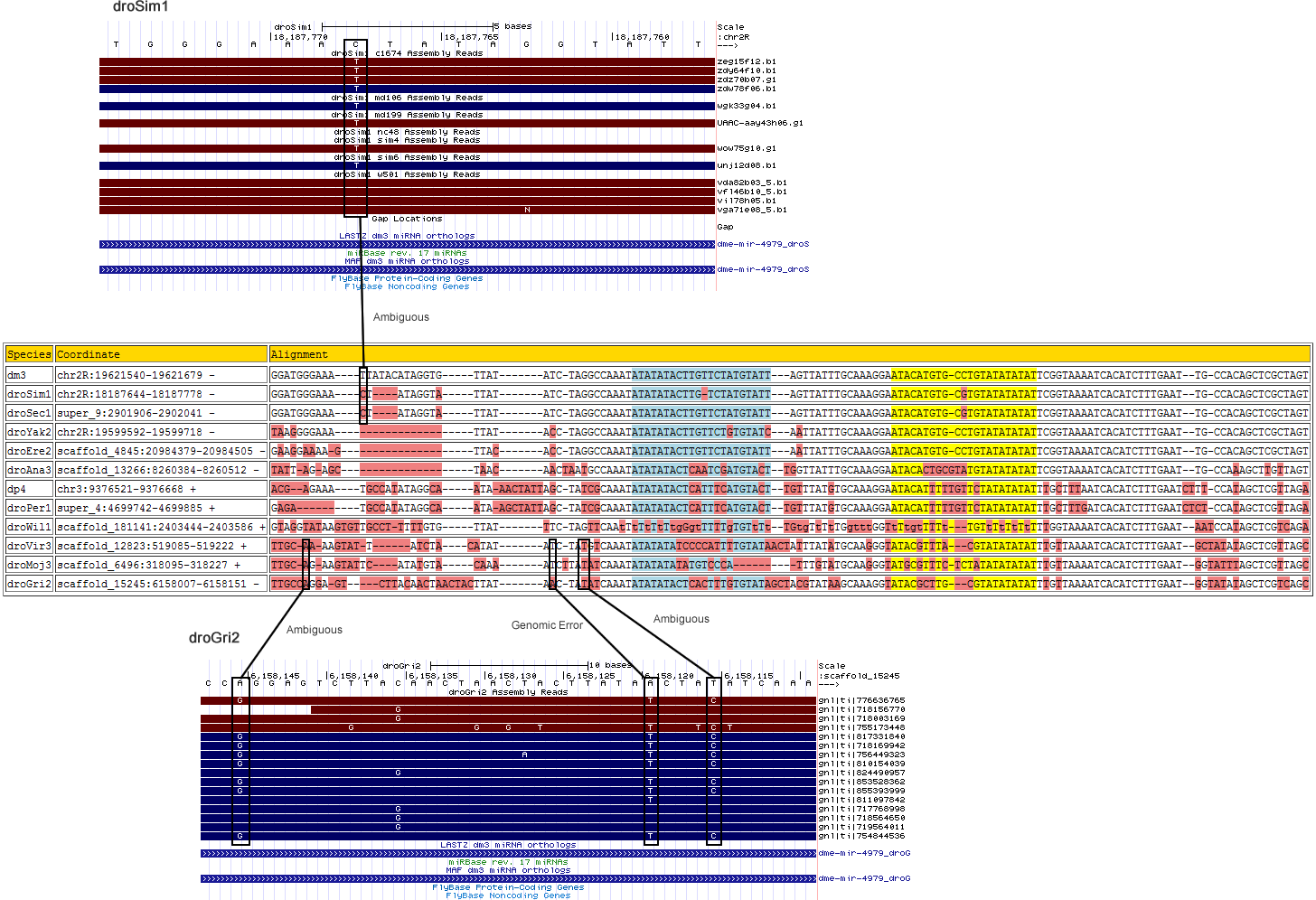
dme-mir-4979
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droGri2 | scaffold_15245 | 6158116 | A | 3 | 0 | 0 | 0 | 0 | 0 | 0.438 | 0 | 0.563 | 0 | 16 | Ambiguous - Suggested base change does not agree with neighboring species |
3' flanking region |
| droGri2 | scaffold_15245 | 6158120 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.625 | 0 | 0 | 0.375 | 16 | Valid - Genomic Error | 3' flanking region |
| droGri2 | scaffold_15245 | 6158146 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.533 | 0 | 0.467 | 15 | Ambiguous - Suggested base change does not agree with neighboring species |
3' flanking region |
| droSim1 | chr2R | 18187768 | G | 3 | 0 | 0 | 0 | 0 | 0 | 0.667 | 0 | 0.333 | 0 | 12 | Ambiguous - The two alleles agree with either melanogaster or sechellia. |
3' flanking region |

Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:19621540-19621679 - | GG-AT----GGGAA--A-------TTATACATAGGTG------TTATATCT-AGGCCAAATATATATACTTG--TTCTATGTA---TTAGTTATTTGCAAAGGAATACATGTG-CCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT |
| droSim1 | chr2R:18187644-18187778 - | GG-AT----GGGAA--A-------CT----ATAGGTA------TTATATCT-AGGCCAAATATATATACTTG--T-CTATGTA---TTAGTTATTTGCAAAGGAATACATGTG-CGTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT |
| droSec1 | super_9:2901906-2902041 - | GG-AT----GGGAA--A-------CT----ATAGGTA------TTATATCT-AGGCCAAATATATATACTTG--TTCTATGTA---TTAGTTATTTGCAAAGGAATACATGTG-CGTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT |
| droYak2 | chr2R:19599592-19599718 - | TA-AG----GGGAA--A--------------------------TTATACCT-AGGCCAAATATATATACTTG--TTCTGTGTA---TCAATTATTTGCAAAGGAATACATGTG-CCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT |
| droEre2 | scaffold_4845:20984379-20984510 - | CT-GTAGAAGGAAA--A------------------------G-TTACACCT-AGGCCAAATATATATACTTG--TTCTATGTA---TTAATTATTTGCAAAGGAATACATGTG-CCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT |
| droAna3 | scaffold_13266:8260384-8260523 - | GG-TT----CAATG--A-------TTAT---TAGA------GCTAACAACTAATGCCAAATATATATACTCA--ATCGATGTA--CTTGGTTATTTGCAAAGGAATACACTGCGTATGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCAAAGCTTGTTAGT |
| dp4 | chr3:9376521-9376668 + | AC-------GAGAA--A-------TGCCATATAGGCAATAAACTATTAGCT-ATCGCAAATATATATACTCA--TTTCATGTA--CTTGTTTATGTGCAAAGGAATACATTTTTGTTCTATATATATTTGCTTTAATCACATCTTTGAATCTTT-CCATAGCTCGTTAGA |
| droPer1 | super_4:4699742-4699885 + | GAGAT----GCCA----------------TATAGGCAATAAGCTATTAGCT-ATCGCAAATATATATACTCA--TTTCATGTA--CTTGTTTATGTGCAAAGGAATACATTTTTGTTCTATATATATTTGCTTTGATCACATCTTTGAATCTCT-CCATAGCTCGTTAGA |
| droWil1 | scaffold_181141:2403442-2403586 + | GT-GT----AGGTATAAGTGTTGCCT----TTTTGTG------TTATTTCT-AGTTCAAATATATATACGCA--TTTTCTGTA--TATGACTATATGCAAAGGTATACATTTA---TGTATATATATTTGGTAAAATCACATCTTTGAAT--AATCCATAGCTCGTCAGA |
| droVir3 | scaffold_12823:519093-519222 + | GT-AT----TAT------------CT----AC-----------ATATATCT-ATGTCAAATATATATATCCCCATTTTGTATA--ACTATTTATATGCAAGGGTATACGTTTA---CGTATATATATTTGTTAAAATCACATCTTTGAAT--GCTATATAGCTCGTTAGC |
| droMoj3 | scaffold_6496:318093-318227 + | AG-TT----GCAGA--A-------GTATTCATATGTA------CAAAATCTTATATCAAATATATATATATG--TCCCA----------TTTGTATGCAAGGGTATGCGTTTC-TCTATATATATATTTGTTAAAATCACATCTTTGAAT--GGTATTTAGCTCGTTAGC |
| droGri2 | scaffold_15245:6158007-6158151 - | TT-GC----CAGGA--G-------TC-T-TACAACTAACTAC-TTATATCT-ATATCAAATATATATACTCA--CTTTGTGTATAGCTACGTATAAGCAAAGGTATACGCTTG---CGTATATATATTTGTTAAAATCACATCTTTGAAT--GGTATATAGCTCGTCAGC |