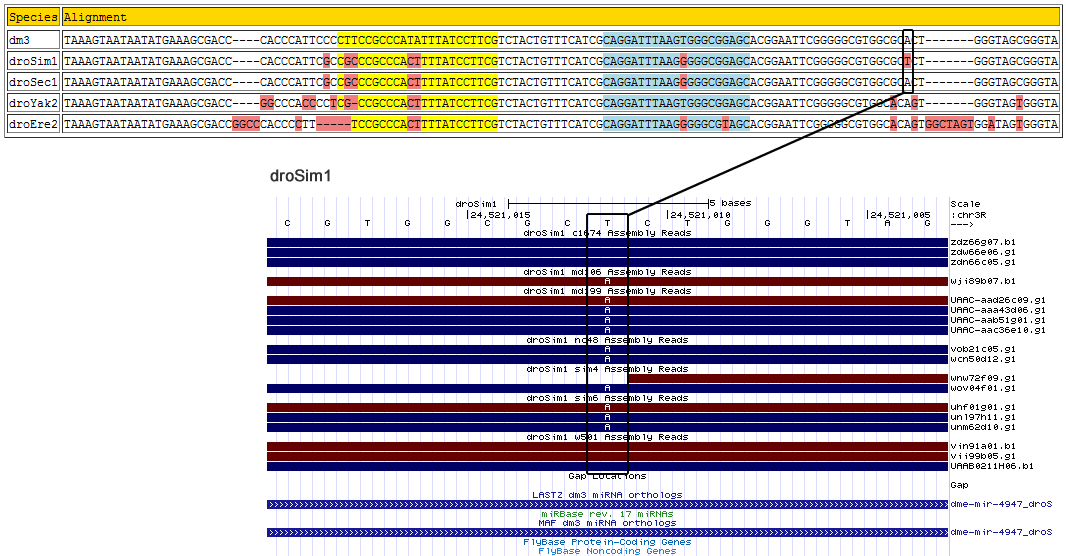
dme-mir-4947
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr3R | 24521012 | A | 3 | 0 | 0 | 0 | 0 | 0 | 0.353 | 0 | 0 | 0.647 | 17 | Genomic Error | 5' flanking region |

Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:24834459-24834589 - | TAAAGTAATAATATGAAAGCGACCCACCCAT--TC------------------------------------------------CCCTTCCGCCCATATTTATCCTT------------------------CGTCTACTGTTTCATCG----CAGGATTTAAGTGGGCGGAGCACGGAAT-----------TCGGGGGCGTGGCGC-----A--CTGGGTAGCGGGTA |
| droSim1 | chr3R:24520998-24521128 - | TAAAGTAATAATATGAAAGCGACCCACCCAT--TC------------------------------------------------GCCGCCCGCCCACTTTTATCCTT------------------------CGTCTACTGTTTCATCG----CAGGATTTAAGGGGGCGGAGCACGGAAT-----------TCGGGGGCGTGGCGC-----A--CTGGGTAGCGGGTA |
| droSec1 | super_4:3696490-3696620 - | TAAAGTAATAATATGAAAGCGACCCACCCAT--TC------------------------------------------------GCCGCCCGCCCACTTTTATCCTT------------------------CGTCTACTGTTTCATCG----CAGGATTTAAGGGGGCGGAGCACGGAAT-----------TCGGGGGCGTGGCGC-----A--CTGGGTAGCGGGTA |
| droYak2 | chr3R:21664594-21664723 + | TAAAGTAATAATATGAAAGCGACCGGCCCAC--C-------------------------------------------------CCTCGCCGCCCACTTTTATCCTT------------------------CGTCTACTGTTTCATCG----CAGGATTTAAGTGGGCGGAGCACGGAAT-----------TCGGGGGCGTGGCAC-----A--GTGGGTAGTGGGTA |
| droEre2 | scaffold_4820:3199292-3199428 + | TAAAGTAATAATATGAAAGCGACCGGCCCAC--C-------------------------------------------------CCTTTCCGCCCACTTTTATCCTT------------------------CGTCTACTGTTTCATCG----CAGGATTTAAGGGGGCGTAGCACGGAAT-----------TCGGGGGCGTGGCACAGTGGCTAGTGGATAGTGGGTA |
| droAna3 | scaffold_13340:19733656-19733776 + | TAAAGTAATAATATAAAAGCGACCAGCCCCT--AC---------------TTCTACTCCACCCAACCA-------------------GCCTCCCGATCCTGGGCAG------------------------CGTTGGTTGTTTCAGAG----G--------------CGGCTTGAGGAAC-----------TCGGGGGCGTGCCGC-----T------------CCTA |
| dp4 | chr3:722533-722667 - | TAAAGAGATAATGACAAAGCAACCCATTAAAAGCCATCCCTTGACAACACTCCTCTCTCATCCC-------------------------------------------------------------------CTTCATATTCCCATCT----CCGAA---AAATGAGCGAAGAACGGAGA-----------GCAACGTTTTAATAA-----A--TTTGTAAACAGATT |
| droPer1 | super_26:1114277-1114391 - | GACGGTGGCCTTCC-------ACTTCCTCTT--CC------------------GCCGCCACCACACCCCCTTCCCACTGCCAA---------GC------A---CG------------------------CTCTTTTTATCGCAATT----G-------TTGTCGG-------CGGA------------------TTTTCGGCTT-----A--CATGCGAGGAGCTA |
| droWil1 | scaffold_181096:10088805-10088934 + | CAAAATGTCCTTGATG-----ACTGGC-------A-------------------------TTCACACACACACA---------CACACACACACACATTTAAACACATCCATATGTACATCCATGTATACATTTATCTAATACAGAT----G--------------CAGAGCTCGGA--------------------------AC-----T--TCAAGTATAAGGCA |
| droVir3 | scaffold_13049:15807-15938 + | TATG-TGGCAGTAGAAATTTCTCCCATAAATATAT-----------------------------GTAC---------------CCTTTCCGTCTTTATGTTTGTTT------------------------GGTTGAATGGATCGAAC----TACAAATCTAGAGATTAGAAATTGGAAA-----------TCCGTATTCTAGTA-----------ATGTATCCAGAA |
| droMoj3 | scaffold_6500:17797597-17797712 + | CACACACAC--------------ACACACAG--AC------------------------------------------------TTTGGCTACAAAATGTTAACTTG------------------------AGTTTGAAATTTTCCAG-----GGAATATGAGTGAGAGGCGAGCGAAAG-----------CAGTGAACAGAGTGA-----A--CAATGTATAAAGTA |
| droGri2 | scaffold_14853:8203259-8203387 + | CAAATTAACAACAAGAATTCTTTCCAGCTCC--CT-------------------------CTCTCTCTCTCTCTCTCTCTCTA---------AC----------TC------------------------CGTCTC-----TCATTTCCGAGAGAGAGCTAGTTACATTGCTGAGAGAGAAGTGAAGAGAGCCGAGC-----------------------TTTGTTA |