dme-mir-4946
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr2L | 18557396 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.588 | 0 | 0 | 0.412 | 17 | Genomic Error | Loop |
| droSim1 | chr2L | 18557397 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.625 | 0 | 0.375 | 16 | Genomic Error | Loop |

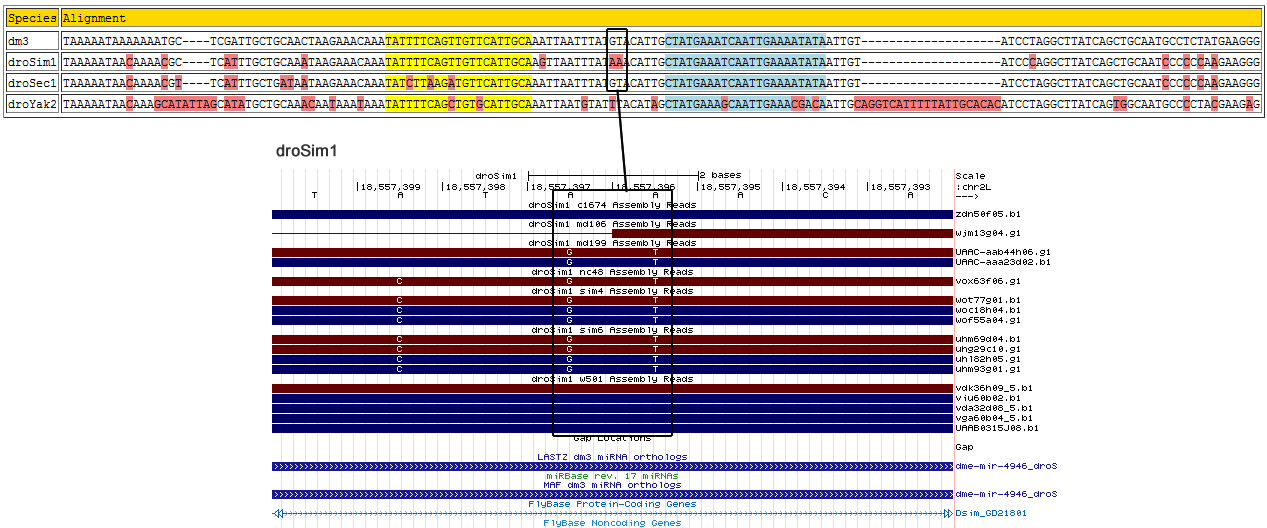
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:18867030-18867176 - | TAAAAATAAAA----AAATG-------------CTC-----------------GATTGCTGCAA-----------------------------------------CTAAGAAACA--AATATTTTCAGTTGTTCAT----TGCAAATTAATTTA---TGTACATTGCTAT---------------GAAATCAATTGAAAATATAAT---TGTA--------------------TCCTAGGCTTATCAGCT-GCAATGCCTCTATGAAGGG |
| droSim1 | chr2L:18557325-18557471 - | TAAAAATAACA----AAACG-------------CTC-----------------ATTTGCTGCAA-----------------------------------------ATAAGAAACA--AATATTTTCAGTTGTTCAT----TGCAAGTTAATTTA---TGTACATTGCTAT---------------GAAATCAATTGAAAATATAAT---TGTA--------------------TCCCAGGCTTATCAGCT-GCAATCCCCCCAAGAAGGG |
| droSec1 | super_7:2494148-2494294 - | TAAAAATAACA----AAACG-------------TTC-----------------ATTTGCTGATA-----------------------------------------ATAAGAAACA--AATATCTTAAGATGTTCAT----TGCAAATTAATTTA---TGTACATTGCTAT---------------GAAATCAATTGAAAATATAAT---TGTA--------------------TCCTAGGCTTATCAGCT-GCAATCCCCCCAAGAAGGG |
| droYak2 | chr2R:5382904-5383074 - | TAAAAATAACA----AAGCA-------------TAT-------------TAGCATATGCTGCAA-----------------------------------------ACAATAAATA--AATATTTTCAGCTGTGCAT----TGCAAATTAATGTA---TTTACATAGCTAT---------------GAAAGCAATTGAAACGACAAT---TGCAGGTCATTTTTATTGCACACATCCTAGGCTTATCAGTG-GCAATGCCCCTACGAAGAG |
| droEre2 | scaffold_4845:5409251-5409398 + | TACAAGTAACA----AAGTG-------------CTA-----------------ATTCGCTGCAA-----------------------------------------ATAGGAAATA--AATATTTTCAT------------------------TATTAT-CACATTGCCA----------------GAAACCAATTGAAAAGATAAT---TGTAGGCCACTTTTGTTGCACGCATCCTAGGCTTATCAGCT-GATATGCCCCCACGAAGGG |
| droAna3 | scaffold_13334:1096999-1097137 - | TAAATATTTTT----AAGTA-------------ATT-----------------AATTG-----------GGCTACTG----------------------------GCTTTAAAAAATAATATTTTCAAATGTTATT----GGTAATTTAATTTT---TTTATAATTTT-TAAA------------TACATTTTTTTAAACTAAAATCCTTGTT--------------------TTCT----------------GCTTCCTTT--TCAGCT |
| dp4 | chrXR_group6:8932576-8932713 + | TTAAAATAAAA----ACATT--------------TC-----------------ATATTTAAATT-ACAAGGTTTTAT----------------------------ATTTTAAATA--AATAT------------ATTTCCCACAAATTAAAGAT---AATATATTTATTT---------------TAAAGTAATTTATAATTAAAT---TGAA--------------------TATGGGCCCTA----C---------TACAAAGACTTT |
| droPer1 | super_1:5889084-5889218 + | GATAGGTT---------------------------T-----------------TTGCAACACAA-----------------------------------------AAAATAAACA--AAGATTAGTTTCCAGTTAT----TAAATATTAATATA---TATATTTGAAGCA---------------GAAATATGTATATTTTAGAAT---CGTT--------------------TCTGTGGGGTTTTCATT-GCAATACCTTT--TATGAA |
| droWil1 | scaffold_180698:8557325-8557469 - | ACCAAAGAGAA----ATATCTCAAACTCAAACTCACAAACAAGACACATCAGCGC--------------------------------GCAAGAGCAACAACGCAAAGAACAAT--------------------------------------------TAAACAATTCTAT---------------TAAATTGATCGAAAATATAAA---CATT--------------------TCA-CAGCATAACAACA-ACAAA-----AGAAAAGGA |
| droVir3 | scaffold_12855:7203873-7204022 + | TGAAAATGAAT----AAATA-------------TAT-------------------ATGCTCCAATTAAAATTGAAACTACTATCCGAT--------------------------------------CGAAGGTCTT-TG-TTCAAATTCATTTG---A--GGGGTTTTTC---------------GCACTTTTTGGAAAATCCATC---ACTC--------------------CCCGATGGAAATGGGCT-TCAATGACTTAGTGGGTGG |
| droMoj3 | scaffold_6500:29247639-29247782 - | GGCCAATTAAC----AAACG-------------CAC-------------------GTGCAGCAG-----------------------------------------CTACAACAGC--ATCGTGTGCACACATACAT----TCAAACT-TCTTCT---GGAACATTCTTTTAATCACATCAGCTGTTAGTTTGGTTTAGTTTTGGTA---GATG--------------------G---------CGCTGCA-G------CACTGAATCTAT |
| droGri2 | scaffold_14853:9426729-9426870 + | TCCACAAAGATATCAAAATC--------------TC-----------------AGATACTGCTT-GAAAGTTGACAT---------------------------------------------TT-----CCCTCAT----AGAATA-TCCTTTA---TGTACAGTAATAT---------------GAGT--GTTTGTATTTGAAGTCT-TCTT--------------------TTTCATCGTCATTTATCTCCATCCTCTCTTTGACTGC |