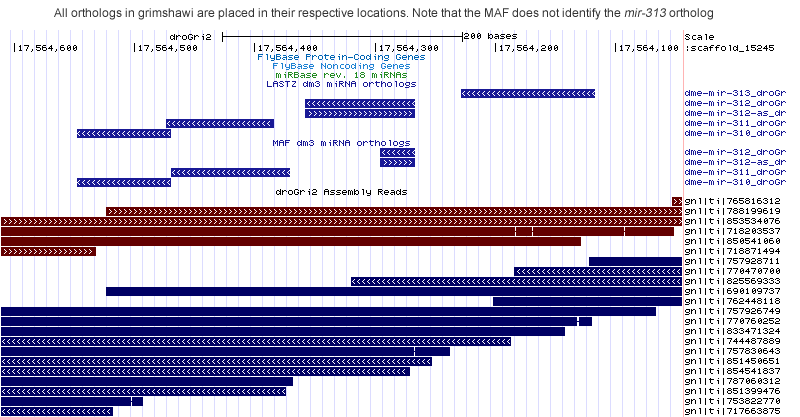
dme-mir-311

Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droGri2 | scaffold_15245 | 17564435 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.714 | 0 | 0 | 0.286 | 14 | Genomic Error | miR |
| droSim1 | chr2R | 15132728 | A | 3 | 0 | 0 | 0 | 0 | 0 | 0.4 | 0 | 0.6 | 0 | 15 | Genomic Error | loop |
| droSim1 | chr2R | 15132760 | G | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.357 | 0.643 | 14 | Genomic Error | 3' flanking region |
The reported D. persimilis ortholog reported in the 12-way alignment is wrong as shown below:
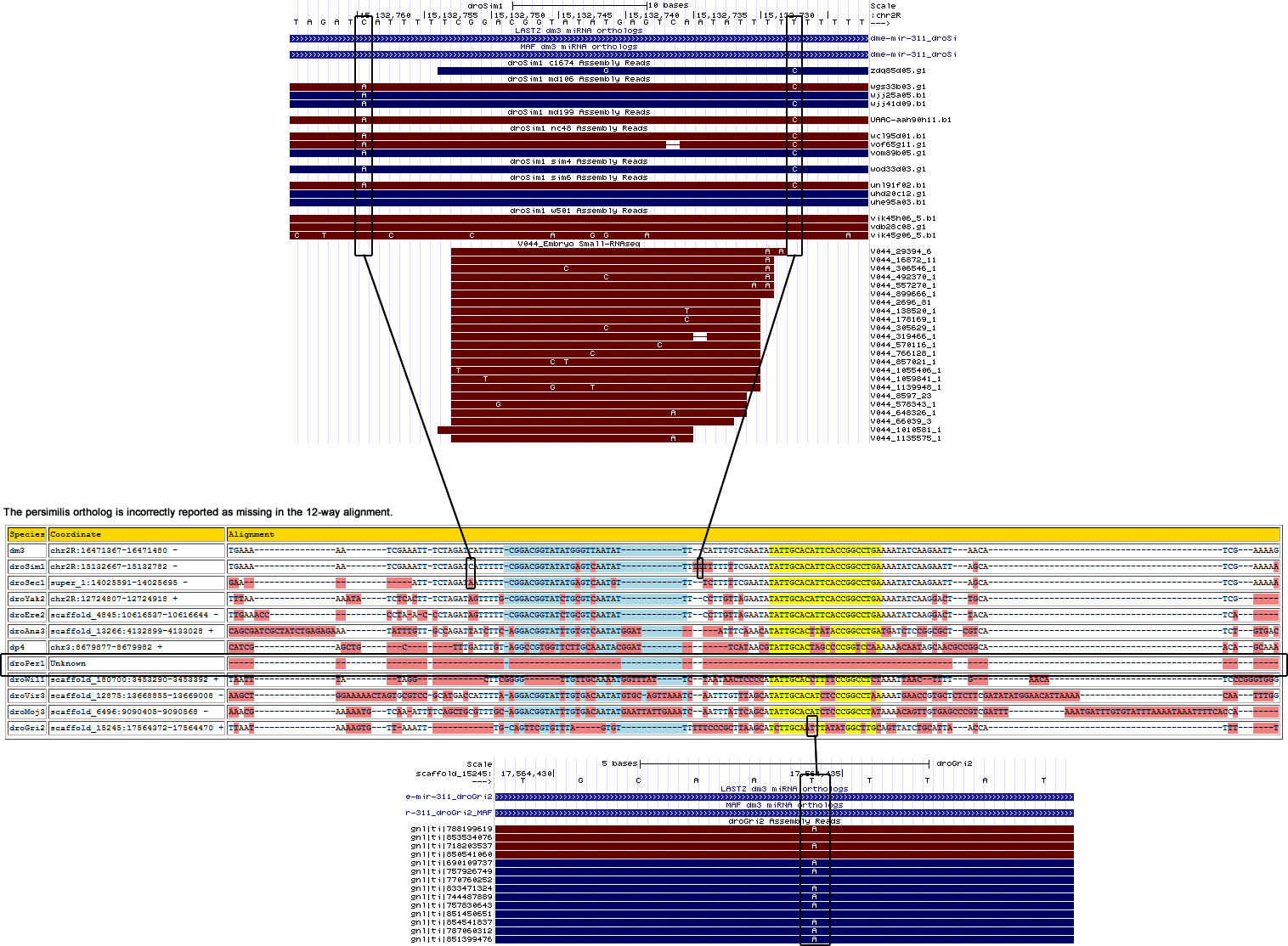
The D. persimilis ortholog is found by searching the persimilis trace data with the D. pseudoobscura ortholog
>dps-mir-311
CATCGAGCTGCTTTGATTTGTAGGCCGTGGTTCTTGCAAATACGGATTCATAACGTATTGCACTAGCCCCGGTCCAAAAAACAATAGCAACGCCGGCAACAGCAAA
>gnl|ti|732560583:591-697
CATCGAGCTGCTTTGATTTGTAGGCCGTGGTTCTTGCAAATACGAATTCATAACGTATTGCACTAGCCCCGGTCCAAAAAACAATAGCAACGCCGGCAACAGCAAA
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471367-16471480 - | TGAAAA-----------------A-TCG---AAATTTCTAGATCATTTTTCGGA--CGGTATATGGGTTAATATTTCAT------------TTGTCGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GAATT-------AACATC-----------------------------------GAA---AAG |
| droSim1 | chr2R:15132667-15132782 - | TGAAAA-----------------A-TCG---AAATTTCTAGATAATTTTTCGGA--CGGTATATGAGTCAATATTTTCT----------TTTTTTCGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GAATT-------AGCATC-----------------------------------GAA---AAA |
| droSec1 | super_1:14025591-14025696 - | GAAA------------------------------TTTCTAGATAATTTTTCGGA--CGGTATATGAGTCAATGTTTTCT------------TTTTCGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GAATT-------AGCATC-----------------------------------GAA---AAA |
| droYak2 | chr2R:12724807-12724923 + | TTTAAA-----------------AA-TATCTCACTTTCTAGATAGTTTTGCGGA--CGGTATCTGCGTCAATATTTCCT------------TGTTAGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GGACT-------TGCATC-----------------------------------GAA---AAG |
| droEre2 | scaffold_4845:10616532-10616644 - | TTGAAA-----------------C-CCC---TA-ACCCTAGATAGTTTTTCGGA--CGGTATCTGCGTCAATATTTCCT------------TGTTAGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GGACT-------TACATC-----------------------------------AAA---AAG |
| droAna3 | scaffold_13266:4132915-4133034 + | AGAGAA-----------------A-TAT---TTGTTGCCAGATTATCTTCAGGA--CGGTATTTGTGTCAATATGGATA-------------TTTCAAACATATTGCACTTATACCGGCCTGATGATCTCC-G--GCGCTCGTCATCTGTGAC-----------------------------------TAA---AAG |
| dp4 | chr3:8679876-8679985 + | CATCGA-----------------GC-TGCTTTGATTTGTA-----------GGC--CGTGGTTCTTGCAAATACGGATT---------------CATAACGTATTGCACTAGCCCCGGTCCAAAAAACAAT-A--GCAA-CGCC---GGCAAC-----------------------------------AGCAAAGTA |
| droPer1 | gnl|ti|732560583:591-697 + | CATCGA-----------------GC-TGCTTTGATTTGTA-----------GGC--CGTGGTTCTTGCAAATACGAATT---------------CATAACGTATTGCACTAGCCCCGGTCCAAAAAACAAT-A--GCAA-CGCC---GGCAAC-----------------------------------AGC---AAA |
| droWil1 | scaffold_180700:3453290-3453392 + | TAAT-------------------------------TTATA-----------GGCTTCGGGGT-TGTTGCAAAATGGTTTATTCT-----AATAACTCCCCATATTGCACCTTTTCCGGCCTCTAAATTAACTTTTG-----------AACATCCCG--------------------------------GGT---GGG |
| droVir3 | scaffold_12875:13668876-13669002 - | GAAAAA-----------------C-TAG-TGCGTCCGCATGACCATTTTAAGGA--CGGTATTTGTGACAATATGTGCAG-TTAAATCAATTTGTTTAGCATATTGCACATCTCCCGGCCTAAAAATGAAC-C--GTGCT-------CTCTTC-----------------------------------GAT---ATA |
| droMoj3 | scaffold_6496:9090405-9090568 - | AAACGAAAAATGTCAAATTTTCAGC-TG---------------CGTTTGCAGGA--CGGTATTTGTGACAATATGAATTATTGAAATCAATTTATTCAGCATATTGCACATCTCCCGGCCTATAAAACAGT-T--GTGA-GCCC---GTCGAT---TTAAATGATTTGTGTATTTAAAATAAATTTTCACC-----A |
| droGri2 | scaffold_15245:17564385-17564474 + | AAAT-------------------------------TTGCAGTTCGTGTTTAG------T-----------GTTTTTTCC------------CGCTTAAGCATCTTGCAAATTATATGGCTTGCAGTTATCT-G--CATTA-------ACCATT-----------------------------------TTC---AGG |