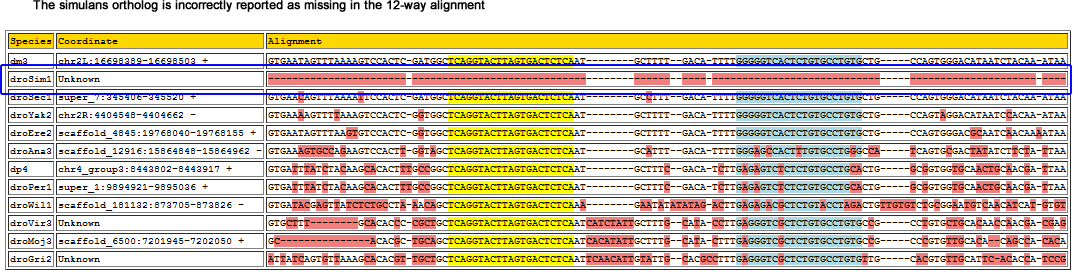
dme-mir-306

The correct ortholog was found in its entirety in 6 of the 7 strains, and partially found in md106.
>dme-mir-306-dm3
GTGAATAGTTTAAAAGTCCACTCGATGGCTCAGGTACTTAGTGACTCTCAATGCTTTTGACATTTTGGGGGTCACTCTGTGCCTGTGCTGCCAGTGGGACATAATCTACAAATAA
>zdv96e12.b1:525-640
GTGAATAGTTTAAAAGTCCACTCGATGGCTCAGGTACTTAGTGACTCTCAATGCCTTTGACATTTTGGGGGTCACTCTGTGCCTGTGCTGCCAGTGGGACATAATCTACAAATAA
This miRNA lie in the same intron of the grp gene as mir-79 and mir-9b, and one intron downstream of mir-9c. The genome browser net track shows that this region's alignment with D. simulans is ambiguous:
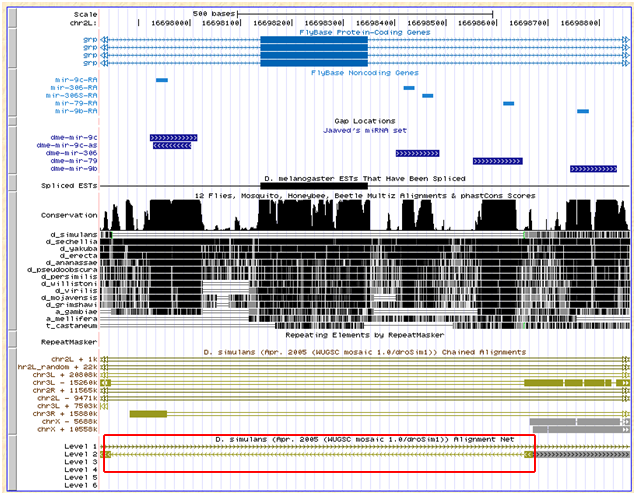
Flybase also does not report an ortholog for grp in D. simulans, however one exists for D. sechellia (GM17194).
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:16698389-16698503 + | GTGAATAG-TTTAAA------AG--------------TCCACTC-G-----ATGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACATA-ATCTACA----AATA-------------A |
| droSim1 | zdv96e12.b1:525-640 + | GTGAATAG-TTTAAA------AG--------------TCCACTC-G-----ATGGCTCAGGTACTTAGTGACTCTCAAT--------GCCTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACATA-ATCTACA----AATA-------------A |
| droSec1 | super_7:345406-345520 + | GTGAACAG-TTTAAA------AT--------------TCCACTC-G-----ATGGCTCAGGTACTTAGTGACTCTCAAT--------GCCTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACATA-ATCTACA----AATA-------------A |
| droYak2 | chr2R:4404548-4404662 - | GTGAAAAG-TTTTAA------AG--------------TCCACTC-G-----GTGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTAGGACATA-ATCCACA----AATA-------------A |
| droEre2 | scaffold_4845:19768040-19768155 + | GTGAATAG-TTTAAG------TG--------------TCCACTC-G-----GTGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACGCA-ATCAACA---AAATA-------------A |
| droAna3 | scaffold_12916:15864848-15864962 - | GTGAAAGT-GCCAGA------AG--------------TCCACTT-G-----GTAGCTCAGGTACTTAGTGACTCTCAAT--------GCATTTGA--CATTTTGGGAGCCACTTTGTGCCTGGGCCA-----TCAGTGCGACTAT-ATCTTCT----ATTA-------------A |
| dp4 | chr4_group3:8443802-8443917 + | GTGATTTA-TCTACA------AG--------------CACACTTTG-----CCGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTCGA--CATCTTGAGAGTCTCTCTGTGCCTGCACTG-----GCGGTGGTGCAAC-TGCAACG----ATTA-------------A |
| droPer1 | super_1:9894921-9895036 + | GTGATTTA-TCTACA------AG--------------CACACTTTG-----CCGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTCGA--CATCTTGAGAGTCTCTCTGTGCCTGCACTG-----GCGGTGGTGCAAC-TGCAACG----ATTA-------------A |
| droWil1 | scaffold_181132:873705-873839 - | GTGACTATACAAAGTGATACGAG--------------TTATCTCTGCCTAAACAGCTCAGGTACTTAGTGACTCTCAAA--------GAATATATATAGACTTGAGAGACGCTCTGTACCTAGACTGTTGTGTCTGCGGAATGTC-AACATCA----TGTG-------------T |
| droVir3 | scaffold_12963:12863192-12863320 - | GTGCTG--------A------ACTC-CTCCCGCCCTGCACACCC-C-----GCTGCTCAGGTACTTAGTGACTCTCAATCATCTATTGCTTTGCA--TACCTTGAGGGTCGCTCTGTGCCTGTGCCG-----CCTGTGCTGCACA-ACCAACG----ACGA-------------G |
| droMoj3 | scaffold_6500:7201931-7202062 + | ATTCATGA-TGTCC-------AG--------------CACACGC-T-----GCAGCTCAGGTACTTAGTGACTCTCAATCACATATTGCTTTGCA--TACTTTGAGGGTCGCTCTGTGCCTGTGCCG-----CCCGTGTTGCACACAGCCACA----CA---ATTGTGA-ATTCG |
| droGri2 | scaffold_14978:830883-831041 + | GTGTTAAA-TGTGGAAAAACCAATCCAATCCG--CTGCACACGT-T-----GCTGCTCAGGTACTTAGTGACTCTCAATTCAACATTGTATTGCA-CGCCTTTGAGGGTCGCTCTGTGCCTGTGTTG-----CACGTGTTGCATT-CACACCATCCGAGTGATTTGTGTCCTTTG |