dme-mir-285
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr3L | 11308082 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.278 | 0 | 0.722 | 18 | Genomic Error | 3' flanking region |
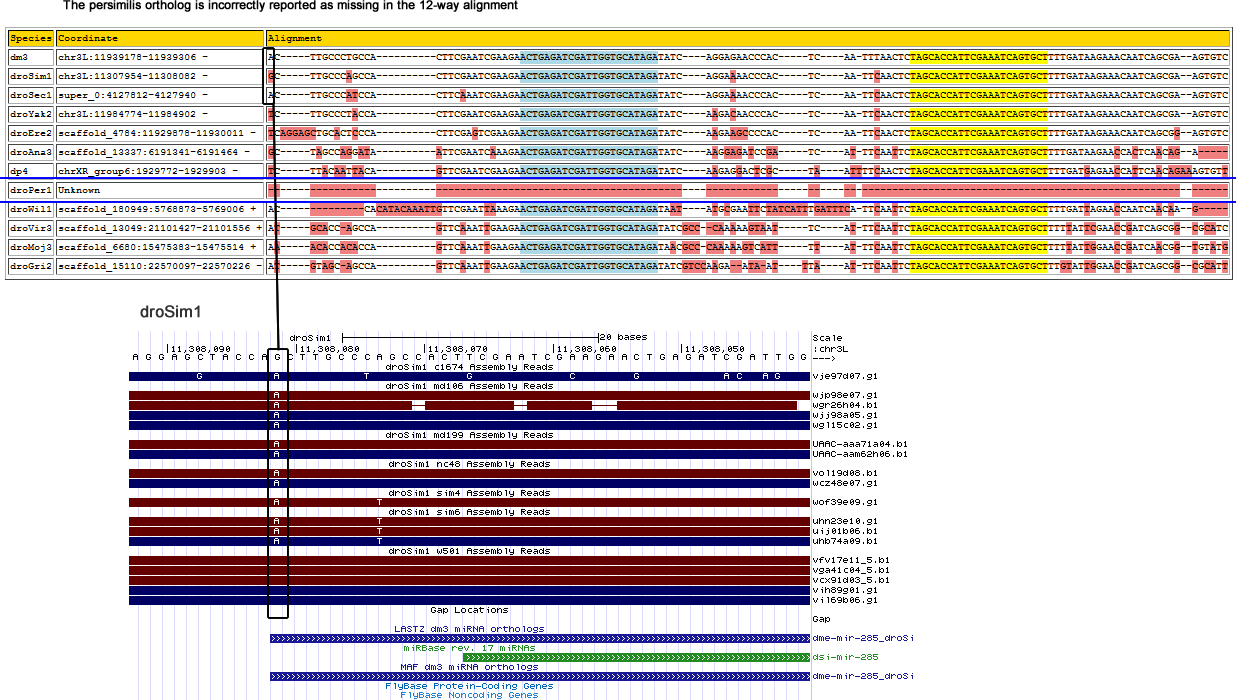
The reported D. persimilis ortholog reported in the 12-way alignment is wrong as shown below:

This miRNA is intergenic and there appear to be no nearby genes. The correct D. persimilis ortholog is found by searching the persimilis trace reads for the D. pseudoobscura sequence via LASTZ.
>dme-mir-285-dp4
TCTTACAAT-TACAGTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATCAAGAGGACTCGCTAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA
>gnl|ti|738578230:636-763
TCTTACAATTTACAGTGGGAATCGAAGAACCTGAGATCGATTGGTGCATAGATATCAAGAGGACTCGATAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:11939178-11939306 - | AC-----TTGCC---CT---------------GCCACTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATC----AGGAGAACCCACTCA---------ATTTAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAA------------GTGTC |
| droSim1 | chr3L:11307954-11308082 - | AC-----TTGCC---CA---------------GCCACTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATC----AGGAAAACCCACTCA---------ATTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAA------------GTGTC |
| droSec1 | super_0:4127812-4127940 - | AC-----TTGCC---CA---------------TCCACTTCAAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATC----AGGAAAACCCACTCA---------ATTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAA------------GTGTC |
| droYak2 | chr3L:11984774-11984902 - | TC-----TTGCC---CT---------------ACCACTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATC----AAGACAACCCACTCA---------ATTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAA------------GTGTC |
| droEre2 | scaffold_4784:11929878-11930011 - | TCAGGAGCTGCA---CT---------------CCCACTTCGAGTCGAAGAAC-TGAGATCGATTGGTGCATAGATATC----AAGAAGCCCCACTCA---------ATTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGGA------------GTGTC |
| droAna3 | scaffold_13337:6191341-6191461 - | GC---------C---AG---------------GATAATTCGAATCAAAGAAC-TGAGATCGATTGGTGCATAGATATC----AAGGAGATCCGATCA---------TTTCAATTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAACCACTCAACAGA----------------- |
| dp4 | chrXR_group6:1929779-1929903 - | TC-----TTACA---AT---------------TACAGTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATC----AAGAGGACTCGCTAAT--------TTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA----------------- |
| droPer1 | gnl|ti|738578230:636-763 + | TC-----TTACA---AT-------------T-TACAGTGGGAATCGAAGAACCTGAGATCGATTGGTGCATAGATATC----AAGAGGACTCGATAAT--------TTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA----------------- |
| droWil1 | scaffold_180949:5768880-5769006 + | AC-----A------------------------AATTGTTCGAATTAAAGAAC-TGAGATCGATTGGTGCATAGATAAT----ATGCGAATTCTATCATTTGATTTCATTCAATTCTAGCACCATTCGAAATCAGTGCTTTTGATTAGAACCAATCAACAAG----------------- |
| droVir3 | scaffold_13049:21101427-21101556 + | AT-----GCAC----CA---------------GCCAGTTCAAATTGAAGAAC-TGAGATCGATTGGTGCATAGATATCGCCCAAAA--AGTAATTCA---------TTTCAATTCTAGCACCATTCGAAATCAGTGCTTTTTATTCGAACCGATCAGCGGC------------GCATC |
| droMoj3 | scaffold_6680:15475365-15475526 + | AC-----ACACCCAGCAGACATCAAACACCACACCAGTTCAAATTGAAGAAC-TGAGATCGATTGGTGCATAGATAACGCCCAAAAAG--TCATTTT----A----TTTCAATTCTAGCACCATTCGAAATCAGTGCTTTTTATTGGAACCGATCAACGGTGTATGGGAACAAGTGTC |
| droGri2 | scaffold_15110:22570097-22570226 - | AT-----GTAG----CA---------------GCCAGTTCAAATTGAAGAAC-TGAGATCGATTGGTGCATAGATATCGTCCAAGAA---TAATTTA----A----TTTCAATTCTAGCACCATTCGAAATCAGTGCTTTGTATTGGAACCGATCAGCGGC------------GCATT |