dme-mir-281-2
Changes

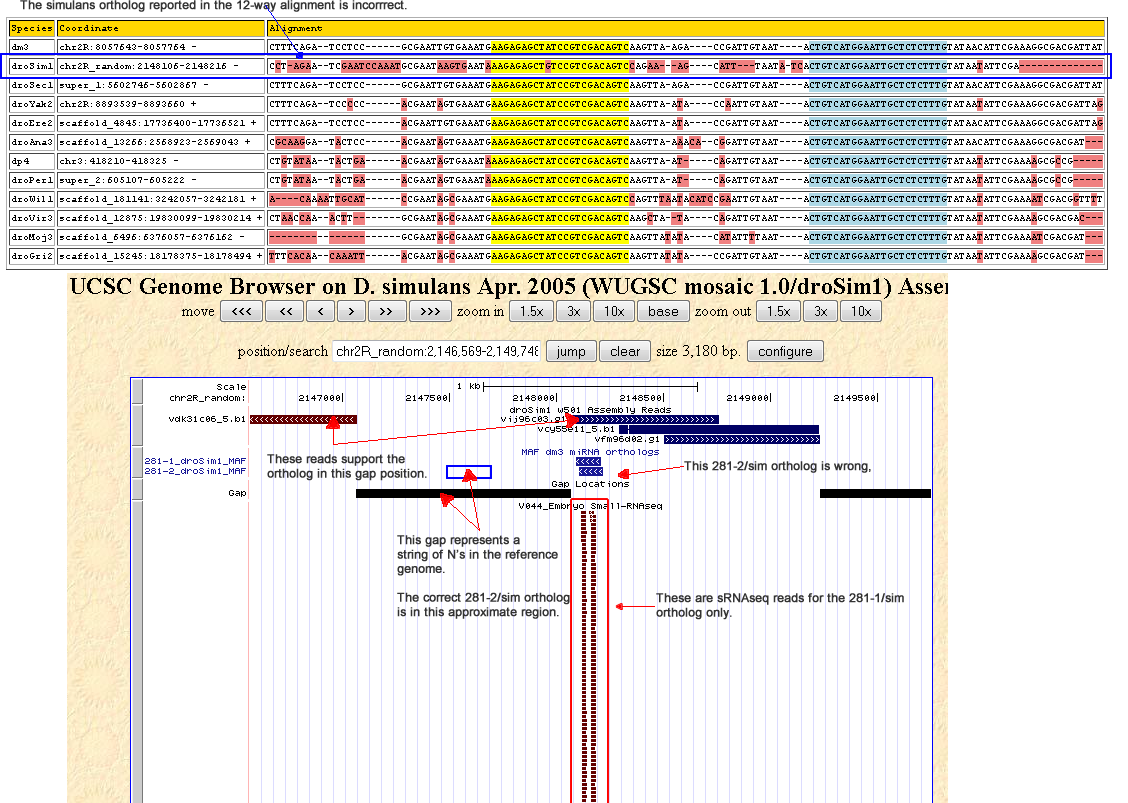
The correct ortholog was fully found in 3 of the 7 simulans strains, and partly found in the remaining 4. This ortholog should lie upstream to the dme-mir-281-1 ortholog, but this region is actually filled by a gap (chr2R_random:2147066-2148065). The MAF actually reports the 281-2 and 281-1 orthologs as the same, but this is clearly not the case apparent from the discordant substitution patterns.
>dme-mir-281-2-dm3
CTTTCAGATCCTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTAAGACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGATTAT
>zdp42d09.g1:294-416
CTTTCAGATCCTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTAAGACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGATTAT
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:8057643-8057764 - | CTTTC-A---G----ATC---CTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA-----AG--ACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGAT--TAT |
| droSim1 | zdp42d09.g1:294-416 + | CTTTC-A---G----ATC---CTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA-----AG--ACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGAT--TAT |
| droSec1 | super_1:5602746-5602867 - | CTTTC-A---G----ATC---CTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA-----AG--ACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGAT--TAT |
| droYak2 | chr2R:8893539-8893660 + | CTTTC-A---G----ATC---CCCCACGAATAGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA-----AT--ACCAATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAGGCGACGAT--TAG |
| droEre2 | scaffold_4845:17736400-17736520 + | CTTTC-A---G----ATC---CTCCACGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA-----AT--ACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGAT--TA- |
| droAna3 | scaffold_13266:2568922-2569043 + | CCGCA-A--GG----ATA---CTCCACGAATAGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA-----AAACACGGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGAT----- |
| dp4 | chr3:418209-418322 - | T-----------ATAATA---CTGAACGAATAGTGAAATAAAGAGAGCTATCCGTCGACAGTCAAGTTA-----A---TCAGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAAGCGCCGA------ |
| droPer1 | super_2:605106-605226 - | TTTTC-T---GTATAATA---CTGAACGAATAGTGAAATAAAGAGAGCTATCCGTCGACAGTCAAGTTA-----A---TCAGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAAGCGCCGA------ |
| droWil1 | scaffold_181141:3242068-3242181 + | TC------------------------CGAATAGCGAAATGAAGAGAGCTATCCGTCGACAGTCCAGTTTAATAC--ATCCGAATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAATCGACGGT--TTT |
| droVir3 | scaffold_12875:19830099-19830219 + | CTAACCAA----------ACT---TGCGAATAGCGAAATGAAGAGAGCTATCCGTCGACAGTCAAGCTA------T--ACAGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAAGCGACGACTTTAT |
| droMoj3 | scaffold_6496:6376053-6376173 - | CCCTC-A---G----------TTCTGCGAATAGCGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA----TAT--ACATATTTTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAATCGACGAT-ATAT |
| droGri2 | scaffold_15245:18178374-18178494 + | GTTTC-A---C---AACAA---ATTACGAATAGCGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTA----TAT--ACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAATATTCGAAAAGCGACGAT----- |