- The correct D. mojavensis ortholog was NOT found by searching the mojavensis trace reads or genome assembly with the orthologs of related species.
- We PCR amplified and resequenced this ortholog successfully.
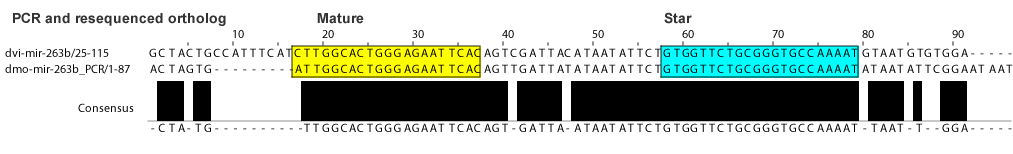
dme-mir-263b

Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr3L | 15181320 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.267 | 0 | 0.467 | 0.267 | 15 | Ambiguous- position appears to be tri-allelic |
5' flanking region |
| droSim1 | chr3L | 15181322 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.733 | 0 | 0.267 | 15 | Genomic Error | 5' flanking region |
| droSim1 | chr3L | 15181375 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0.867 | 0.133 | 0 | 0 | 15 | Ambiguous - The two alleles agree with either melanogaster or sechellia. | Loop |
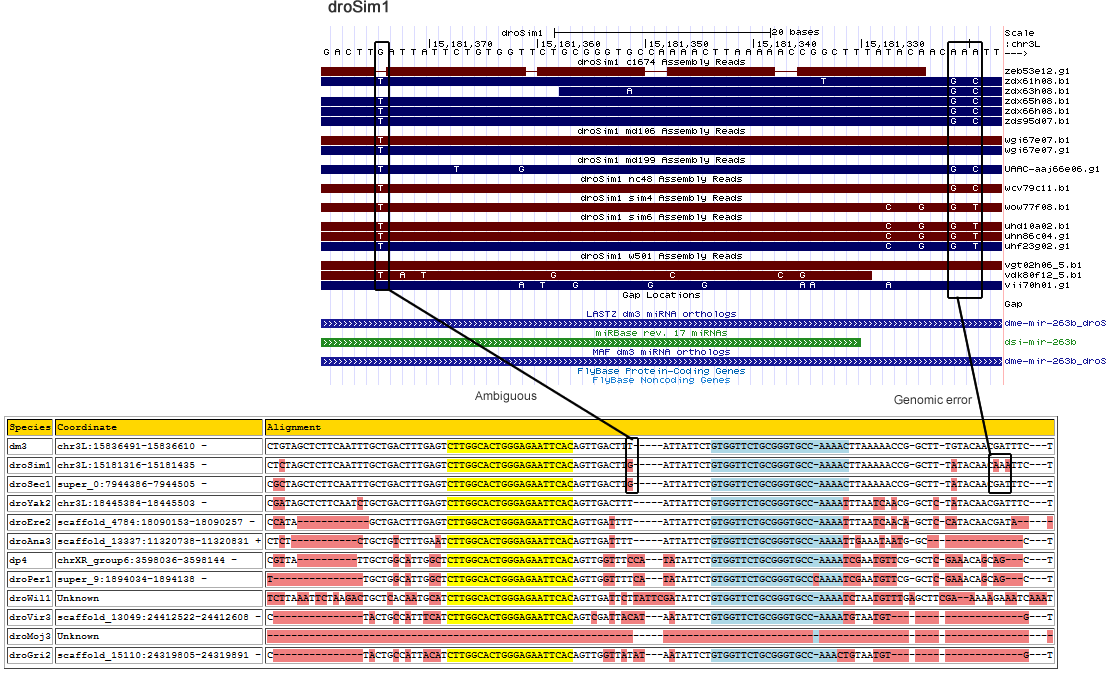
Corrected Alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:15836491-15836610 - | CTGTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTGTACAACGATT------------TCT |
| droSim1 | chr3L:15181316-15181435 - | CTCTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGAAT------------TCT |
| droSec1 | super_0:7944386-7944505 - | CGCTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGATT------------TCT |
| droYak2 | chr3L_random:2539138-2539257 - | CGATAGCTCTTCA-----------------------------A------TCTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACG-GCTCTATACAACGATT------------TCT |
| droEre2 | scaffold_4784:18090156-18090270 - | CTG--GCGATTCA-----------------------------AACC---ATAGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACA-GCTCCATACAACG------------------ |
| droAna3 | scaffold_13337:11320710-11320858 + | TTGTGATTTTTGAATCAAATATTCTG--------------G----ACTCTCTGCTGTCTTTGAATCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTGAAATAATG-GCCTAAAACCTCAAAAAAGGATATTTATTAT |
| dp4 | chrXR_group6:3598033-3598148 - | CGATCGTTA-----------------------------------------TTGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTCCAT---ATATTCTGTGGTTCTGCGGGTGCC-AAAATCGAATGTTCG-GCTCGAAACAGCAGCT------------GAA |
| droPer1 | super_9:1894031-1894138 - | T--------------------------------------------------TGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTTCAT---ATATTCTGTGGTTCTGCGGGTGCCCAAAATCGAATGTTCG-GCTCGAAACAGCAGCT------------GAA |
| droWil1 | scaffold_180727:1452378-1452537 + | CTTAAATTCTAAG-AAAGATTTTCTGTGTGTGTTTGAACGAAAATAAAGTCTGCTCACAATGCATCTTGGCACTGGGAGAATTCACAGTTGATTCTTATTCGATATTCTGTGGTTCTGCGGGTGCC-AAAATCTAATGTTTGAGCTGTGATCATCGAAA------------AAG |
| droVir3 | scaffold_13049:24412507-24412633 - | TTGT-GTTCTTCT------------------------ACGGAAAGCAGAGCTACTGCCATTTCATCTTGGCACTGGGAGAATTCACAGTCGATTACATA---ATATTCTGTGGTTCTGCGGGTGCC-AAAATGTAATGTGTG-GACCAAGACAGC-----------------CT |
| droMoj3 | Unknown | AC-TAGT---------------------------------------------------------GATTGGCACTGGGAGAATTCACAGTTGATTATATA---ATATTCTGTGGTTCTGCGGGTGCC-AAAATATAATATTCG-GAATA------------------------AT |
| droGri2 | scaffold_15110:24319786-24319904 - | ATATTGT--GGAA-----------------------------A------GCTACTGCCATTACATCTTGGCACTGGGAGAATTCACAGTTGGTTATATA---ATATTCTGTGGTTCTGCGGGTGCC-AAACTGTAATGTGTG-GACATAGA-AACAACC------------TGA |