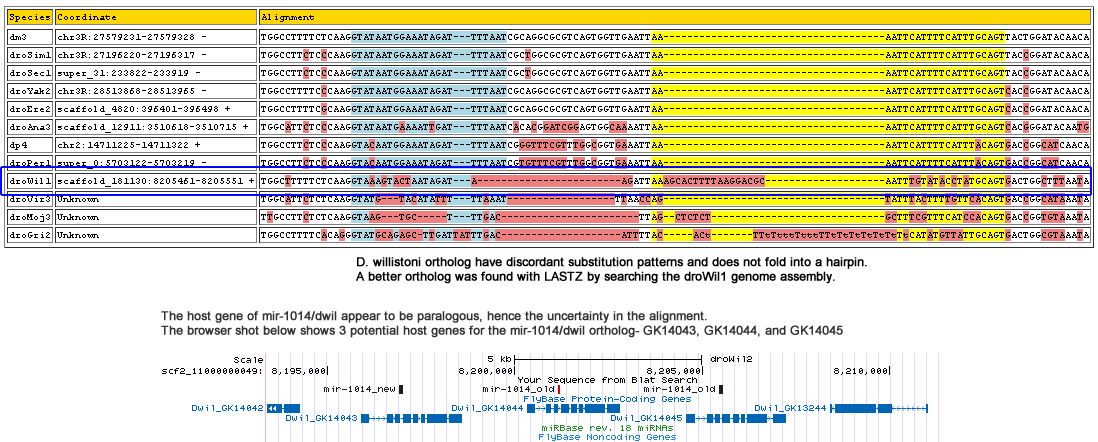
dme-mir-1014

Corrected Alignment:
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:27579231-27579328 - | TGGCCTTTTCTCAAGGTATAATGGAAATAGATTTTAAT------CGCAG----GC---------GCGTC----AGTGG-----TTG--AATTAA------------AATTCATTTTCATTTGCAGTTACTGGATACAACA |
| droSim1 | chr3R:27196220-27196317 - | TGGCCTTCTCCCAAGGTATAATGGAAATAGATTTTAAT------CGCTG----GC---------GCGTC----AGTGG-----TTG--AATTAA------------AATTCATTTTCATTTGCAGTTACCGGATACAACA |
| droSec1 | super_31:233822-233919 - | TGGCCTTCTCCCAAGGTATAATGGAAATAGATTTTAAT------CGCTG----GC---------GCGTC----AGTGG-----TTG--AATTAA------------AATTCATTTTCATTTGCAGTTACCGGATACAACA |
| droYak2 | chr3R:28513868-28513965 - | TGGCCTTTTCCCAAGGTATAATGGAAATAGATTTTAAT------CGCAG----GC---------GCGTC----AGTGG-----TTG--AATTAA------------AATTCATTTTCATTTGCAGTCACCGGATACAACA |
| droEre2 | scaffold_4820:396401-396498 + | TGGCCTTTTCGCAAGGTATAATGGAAATAGATTTTAAT------CGCAG----GC---------GCGTC----AGTGG-----TTG--AATTAA------------AATTCATTTTCATTTGCAGTCACCGGATACAACA |
| droAna3 | scaffold_12911:3510618-3510715 + | TGGCATTCTCCCAAGGTATAATGAAAATTGATTTTAAT------CACAC----GG---------ATCGG----AGTGG-----CAA--AATTAA------------AATTCATTTTCATTTGCAGTCACGGGATACAATG |
| dp4 | chr2:14711225-14711322 + | TGGCCTTCTCCCAAGGTACAATGGAAATAGATTTTAAT------CGGGT----TT---------CGTTT----GGCGG-----TGA--AATTAA------------AATTCATTTTCATTTACAGTGACCGGCATCAACA |
| droPer1 | super_0:5703122-5703219 - | TGGCCTTCTCCCAAGGTACAATGGAAATAGATTTTAAT------CGTGT----TT---------CGTTT----GGCGG-----TGA--AATTAA------------AATTCATTTTCATTTACAGTGACCGGCATCAACA |
| droWil1 | scaffold_181130:8196915-8197021 + | TGGCATTTTCTCAAGGTGAAATGAAAATAAATTTTAATTCGGATAGAAA----GC---------TAATCTAATTGT-A-----TTG--CATTAA------------AATTCGTTTTCATTTGCAGTATCTGGTCTAAATA |
| droVir3 | Unknown | TGGCATTCTCTCAAGGTATG---TACATATTTTTAAAT----------------------------------------------TT--AACCAG------------TATTTACTTTTGTTCACAGTGACCGGCATAAATA |
| droMoj3 | scaffold_6540:30646887-30646982 + | TTGCCTTCTCTCAAGGTAAG--------T-GCTTTGACT-----T---------A---------GCAAA----AGTTA-----TTT---ACTAGAAGTGTTTCTCTGCTTTCGTTTCATCCACAGTGACCGGTGTAAATA |
| droGri2 | scaffold_14624:997050-997159 - | TGGCCTTTTCACAGGGTATGCAGAGCT-TGATTA------------TTTGATTGCTCATTTTACACATT----A-TAAATAAATTATATATATA------------TATACATATGTTATTGCAGTGACTGGCGTAAATA |