| (1) KIDNEY | (1) OTHER | (2) TESTES |
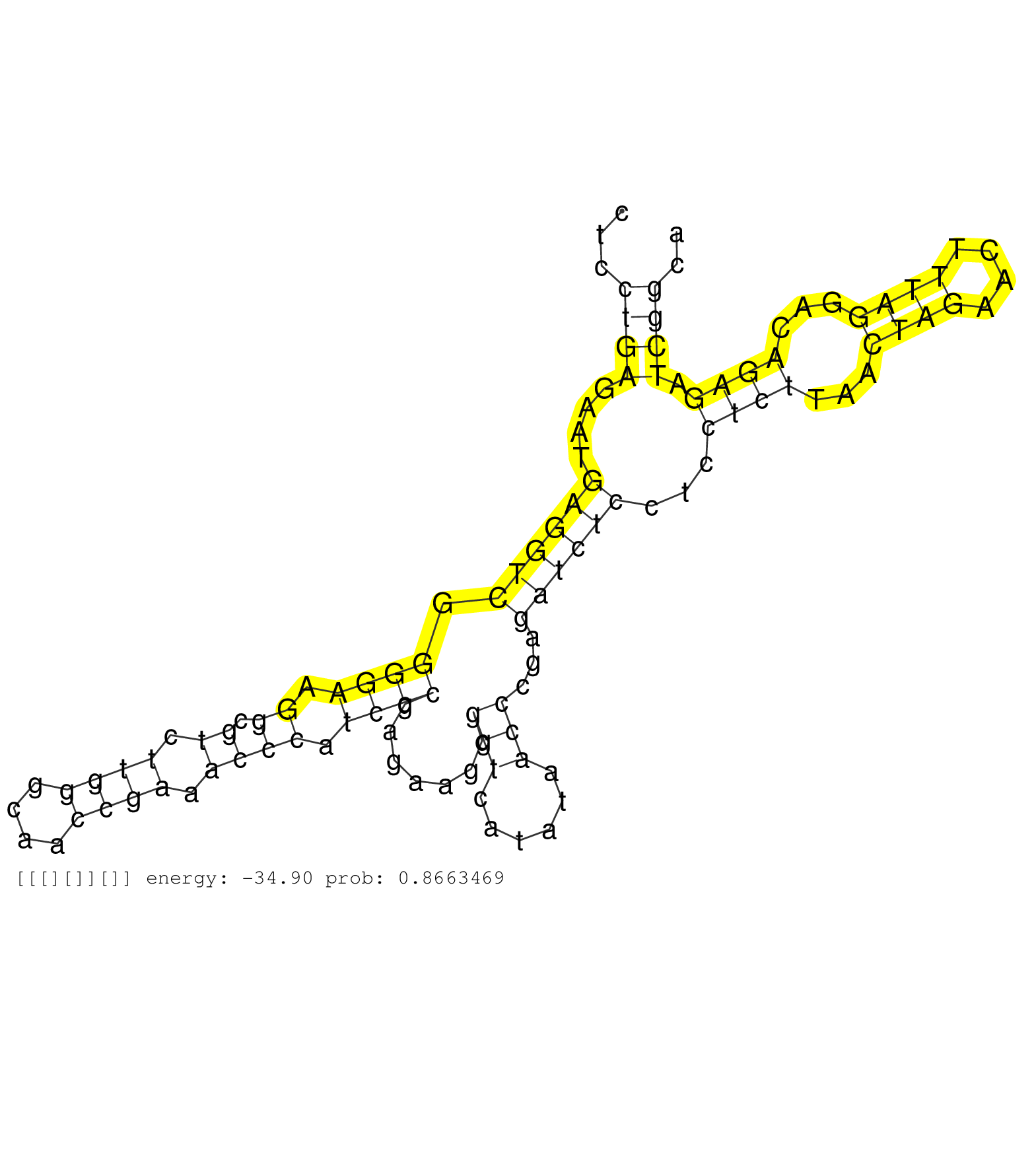
| GTGATTGCAGGGGATTTTCGGTGGACGTTAGGCGACGGAGGGGGGCTCCTGTAAGAGACTGAGGTTTCTGTCCGCAGCACTGCCTTCGCTGAAAAGAGCTTGCATCTGAGCCCCCGCACCGCAGGAGCGAGCCAGGCTCCTGAGAATGAGGTCGGGGAAGGCGTCTTGGGCAACCGAAACCCATCCCGAGAAGCGGTCATATAACCCGAGATCTCCTCCTCTTAACTAGAACTTTAGGACAGAGATCGGCAAAGTCTGAGGAAATGGGCACGGCTTAGAGCTGCCGGCGGCTGCGGAATTTACTTAGCTTCTTGGTTTTTTGGGGGGAAGACTTGGCTTGTTTAATCGAA ...........................................................................................................................................((((....((((((.((((.((.((.((((....)))).)))).)))).......(((......)))...))))))...((((...((((....))))...)))).))))..................................................................................................... ........................................................................................................................................137...............................................................................................................251................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553606(SRX182812) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................TAACTAGAACTTTAGGACAGAGATCGGC.................................................................................................... | 28 | 1 | 7.00 | 7.00 | 5.00 | 2.00 | - | - |
| ..............................................................................................................................................................................................................................TAACTAGAACTTTAGGACAGAGATCGGCA................................................................................................... | 29 | 1 | 3.00 | 3.00 | 3.00 | - | - | - |
| ..............................................................................................................................................................................................................................TAACTAGAACTTTAGGACAGAGATC....................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - |
| .............................................................................................................................................................................................................................TTAACTAGAACTTTAGGACAGAGATCGGC.................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................................................................AACTAGAACTTTAGGACAGAGATCGGCAA.................................................................................................. | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - |
| ..............................................................................................................................................................................................................................TAACTAGAACTTTAGGACAGAGATCGGCAA.................................................................................................. | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - |
| .............................................................................................................................................GAGAATGAGGTCGGGGAAG.............................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................................................GAGACTGAGGTTTCTGTCCGCAGCACTGC........................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................................................TAGGACAGAGATCGGCAAAGTCTGAGGAA....................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................................TAACTAGAACTTTAGGACAGAGATCGG..................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................TAGAACTTTAGGACAGAGATCGGCAA.................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................TCTTAACTAGAACTTTAGGACAGAGATCGGC.................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| GTGATTGCAGGGGATTTTCGGTGGACGTTAGGCGACGGAGGGGGGCTCCTGTAAGAGACTGAGGTTTCTGTCCGCAGCACTGCCTTCGCTGAAAAGAGCTTGCATCTGAGCCCCCGCACCGCAGGAGCGAGCCAGGCTCCTGAGAATGAGGTCGGGGAAGGCGTCTTGGGCAACCGAAACCCATCCCGAGAAGCGGTCATATAACCCGAGATCTCCTCCTCTTAACTAGAACTTTAGGACAGAGATCGGCAAAGTCTGAGGAAATGGGCACGGCTTAGAGCTGCCGGCGGCTGCGGAATTTACTTAGCTTCTTGGTTTTTTGGGGGGAAGACTTGGCTTGTTTAATCGAA ...........................................................................................................................................((((....((((((.((((.((.((.((((....)))).)))).)))).......(((......)))...))))))...((((...((((....))))...)))).))))..................................................................................................... ........................................................................................................................................137...............................................................................................................251................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553606(SRX182812) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ..........................................................gtgGGACAGAAACCTGTG.................................................................................................................................................................................................................................................................................. | 18 | 2.00 | 0.00 | - | - | 2.00 | - | |
| ........................................................................................................................................................CAAGACGCCTTCCCCG...................................................................................................................................................................................... | 16 | 1 | 2.00 | 2.00 | - | - | 2.00 | - |
| ............................................................ccGCGGACAGAAACCCC................................................................................................................................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | 1.00 |