| (1) HEART | (2) KIDNEY | (3) OTHER | (2) TESTES |
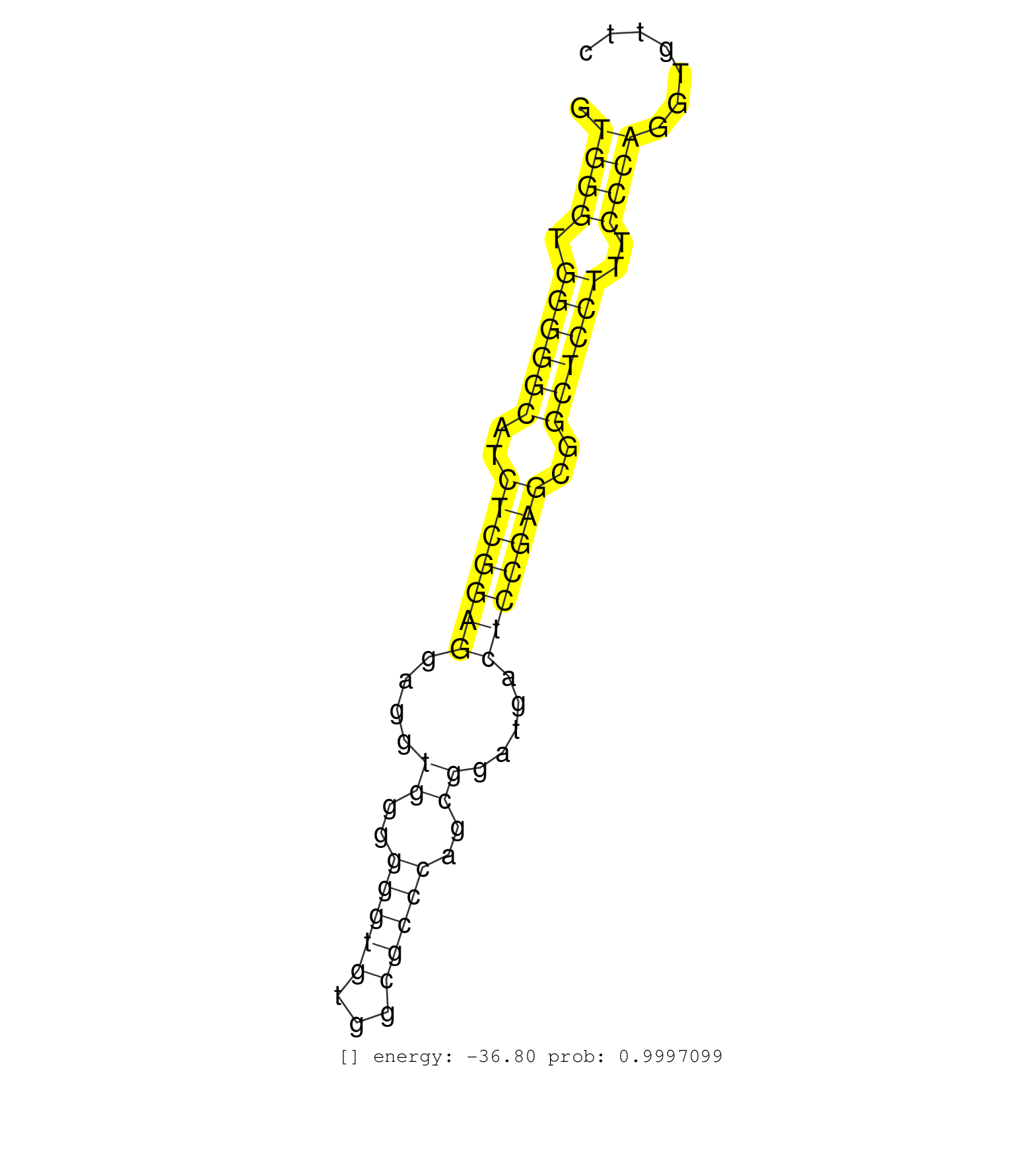
| CTGCCACCGACTGGGATGACAAGAGCATCCGACAGGCCTTCATCCGCAAGGTGGGTGGGGGCATCTCGGAGGAGGTGGGGGGTGTGGCGCCCAGCGGATGACTCCGAGCGGCTCCTTTCCCAGGTGTTCCTGGTGCTGACCCTGCAGCTGTCGGTGACCCTGTCCACGGTGTC ...................................................((((.((((((..(((((((....((..(((((...)))))..)).....)))))))..))))))..))))................................................... ..................................................51............................................................................129.......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .TGCCACCGACTGGGATGACAAGAGC................................................................................................................................................... | 25 | 1 | 6.00 | 6.00 | 5.00 | 1.00 | - | - | - | - | - | - |
| ..................................................GTGGGTGGGGGCATCTCGGAG...................................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - |
| ............................................................................................................................TGTTCCTGGTGCTGACCCTGCAGCTGTCGGT.................. | 31 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................CTGTCGGTGACCCTGTCCACGGTGTC | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| .......................................................................................................CCGAGCGGCTCCTTTCCCAGTT................................................ | 22 | 2.00 | 0.00 | - | - | 1.00 | 1.00 | - | - | - | - | |
| ..................................................GTGGGTGGGGGCATCTCGGA....................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................AGCGGATGACTCCGAGCGGCTCCTTTCCCAGT................................................. | 32 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | |
| .TGCCACCGACTGGGATGACAAGAGCATC................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................ATGACTCCGAGCGGCTCCTT........................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................CCGAGCGGCTCCTTTCCCAGTA................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | |
| .TGCCACCGACTGGGATGACAAGAGCA.................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGGGTGGGGGCATCTCGGAG...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................GCAGCTGTCGGTGACCCTGTCCACGGT... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGCAGCTGTCGGTGACCCTGTCCAC...... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................GCAGCTGTCGGTGACCCTGTCC........ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................TCCGAGCGGCTCCTTTCCCA................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................TGTTCCTGGTGCTGACCCTGCAGCTGTTGGT.................. | 31 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | |
| ................TGACAAGAGCATCCGAC............................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ................TGACAAGAGCATCCGACAGGCCTTCATCC................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................TCCGAGCGGCTCCTTTCCCAGA................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | |
| ...............................................................................................................................TCCTGGTGCTGACCCTGCAGCTGTCGGT.................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................TGTCGGTGACCCTGTCCACGGTGTC | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................TCCGAGCGGCTCCTT........................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - |
| CTGCCACCGACTGGGATGACAAGAGCATCCGACAGGCCTTCATCCGCAAGGTGGGTGGGGGCATCTCGGAGGAGGTGGGGGGTGTGGCGCCCAGCGGATGACTCCGAGCGGCTCCTTTCCCAGGTGTTCCTGGTGCTGACCCTGCAGCTGTCGGTGACCCTGTCCACGGTGTC ...................................................((((.((((((..(((((((....((..(((((...)))))..)).....)))))))..))))))..))))................................................... ..................................................51............................................................................129.......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|