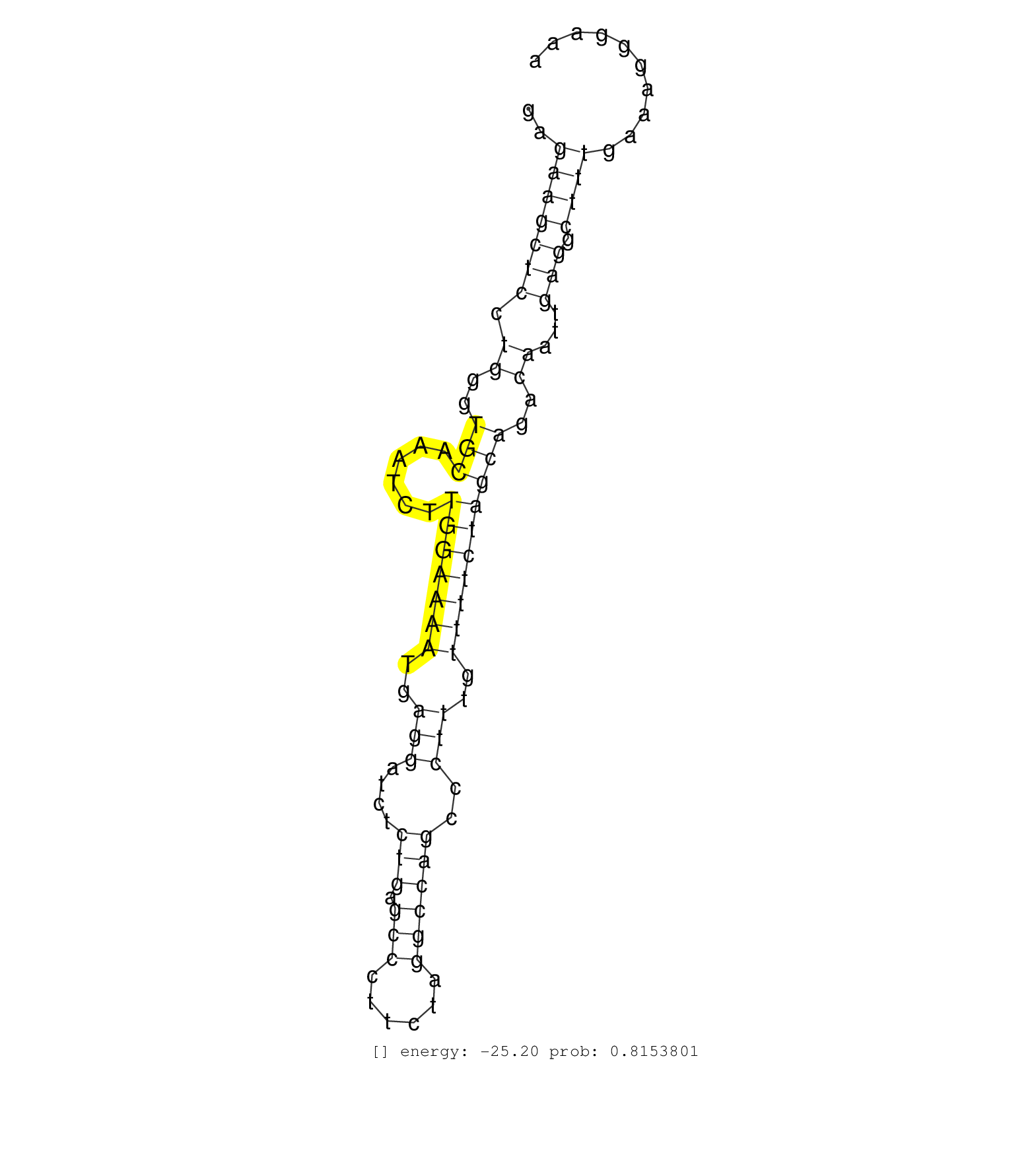
| (1) HEART | (2) TESTES |
| CAAAGGCATTGTAGACATTCTTCCTGCAAGACTTGGGAACCGACAAATAGGTGTGCCCTGCTTGCTAGAGTTTTTGTGCAGGAACAGCCCCTCCCGTCTTTGTCCTGGCCGTGAGCACCAGGGCTAAGCTTTTGAACACTTCCTTTATGTTTGGATTCAGCCCAGGCAATGCATATTTGCTTTCATTTCTTCTTGAGCTTGAGAAGCTCCTGGGTGCAAATCTTGGAAAATGAGGATCTCTGAGCCCTTCTAGGCCAGCCCTTTGTTTTCTAGCAGACAATTGAGGCTTTGAAAGGGAAAGTGGGTGCGGGCACCCCACAGGTGGCCCTCATCACCCAATTGCCAGTGCC ..........................................................................................................................................................................................................(((((((.((..(((......(((((((..(((....(((.(((......))))))..)))..))))))))))..))...))).))))............................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................AATCTTGGAAAATGAGGATCTCTGAGC......................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ......................................................................................................................................................................................................................TGCAAATCTTGGAAAAG....................................................................................................................... | 17 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................................................................................................................................AGAAGCTCCTGGGTGCAGAAA................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | 1.00 | |
| ..........GTAGACATTCTTCCTGCAAGACT............................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..................................................................................................................................TTTGAACACTTCCTTTATGTTTGGATTCAGC............................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ........TTGTAGACATTCTTCCTGCAAGACTTGGGA........................................................................................................................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..........................................................TGCTTGCTAGAGTTTTTGTGCAGGAACAGC...................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................................................................................................ATCTTGGAAAATGAGGATCTCTGAGC......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...AGGCATTGTAGACATTCTTCCTGCA.................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| CAAAGGCATTGTAGACATTCTTCCTGCAAGACTTGGGAACCGACAAATAGGTGTGCCCTGCTTGCTAGAGTTTTTGTGCAGGAACAGCCCCTCCCGTCTTTGTCCTGGCCGTGAGCACCAGGGCTAAGCTTTTGAACACTTCCTTTATGTTTGGATTCAGCCCAGGCAATGCATATTTGCTTTCATTTCTTCTTGAGCTTGAGAAGCTCCTGGGTGCAAATCTTGGAAAATGAGGATCTCTGAGCCCTTCTAGGCCAGCCCTTTGTTTTCTAGCAGACAATTGAGGCTTTGAAAGGGAAAGTGGGTGCGGGCACCCCACAGGTGGCCCTCATCACCCAATTGCCAGTGCC ..........................................................................................................................................................................................................(((((((.((..(((......(((((((..(((....(((.(((......))))))..)))..))))))))))..))...))).))))............................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|