| (2) KIDNEY | (2) OTHER | (2) TESTES |
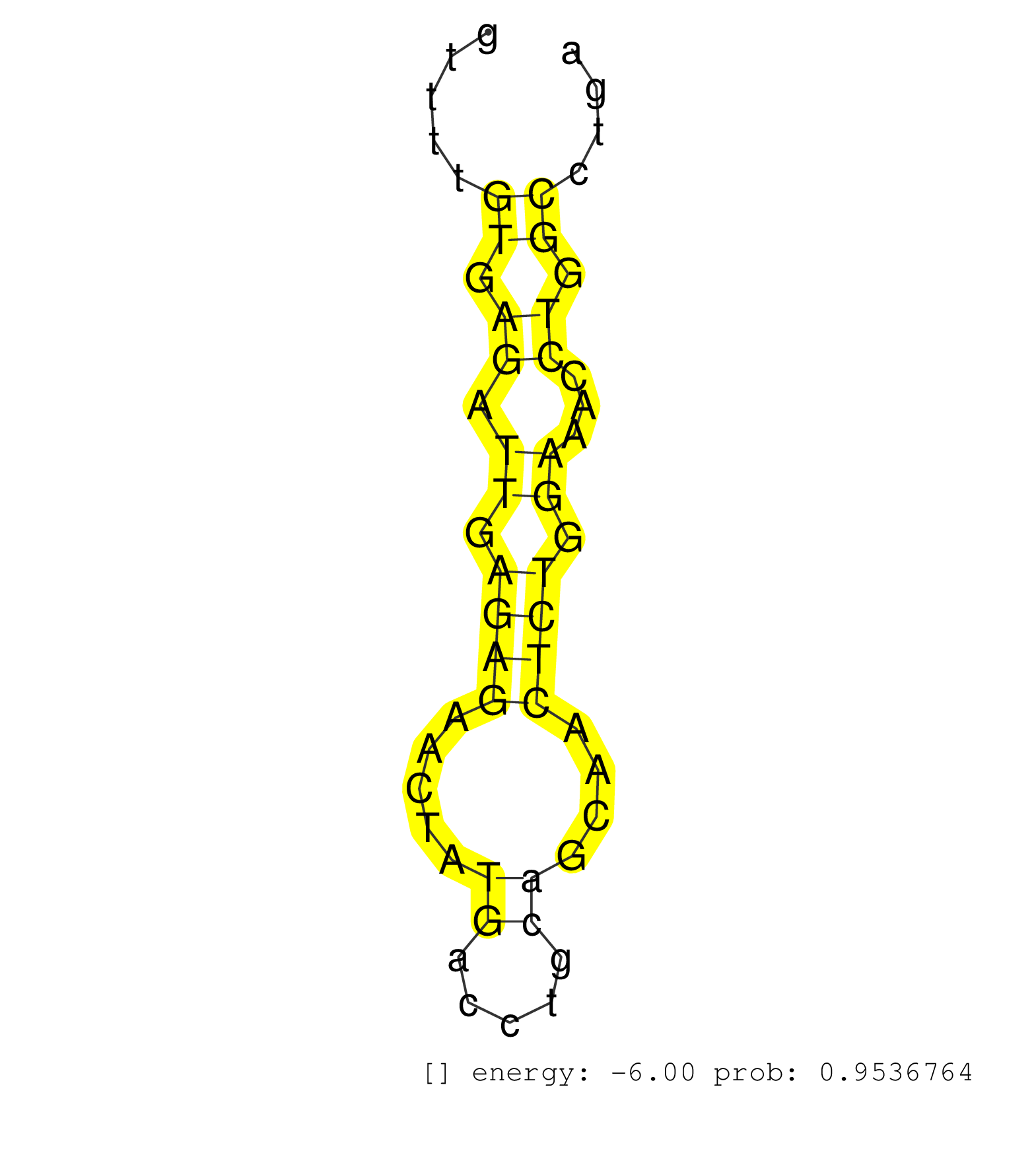
| GCCTACAACAAGCTCCACACATGGCAGTCACGGCGTCTGATGAAGAAAACGTGAGGTGGCCATAATGCTCACAGGTTTTGTGAGATTGAGAGAACTATGACCTGCAGCAACTCTGGAAACCTGGCCTGACAGACAAGCAGATGACCTCACAGGAGTGATAAGAAACATCTGCTTCACGCCAACTCCCAGAGCTGATGCTATTGTACTTGCACATTGGGGACTGAAAGGAAAGAAGGGACTAAATGCTGGGGAGGTAAATTAAGGCAGAACCAAATGAGCTACGTTGCAAATATATATATATACACACATGCACATATATATGTACCTGTGTATGTACATATATATATTTT ...............................................................................((.((.((.((((.....((.....))....)))).))...)).))................................................................................................................................................................................................................................. ..........................................................................75....................................................129........................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553607(SRX182813) source: Testis. (testes) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................GTGAGATTGAGAGAACTATG........................................................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - |
| ............................................................................TTTGTGAGATTGAGAGAACTATGACCTGC..................................................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - |
| ................................................................................TGAGATTGAGAGAACTATGACC........................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - |
| .............................................................................TTGTGAGATTGAGAGAACTA............................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ................................................................................TGAGATTGAGAGAACTATGACCT....................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ................................................................................TGAGATTGAGAGAACTATGACCTGCAGCT................................................................................................................................................................................................................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | |
| .............................................................................TTGTGAGATTGAGAGAACTATGT.......................................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | |
| ..............................................................................TGTGAGATTGAGAGAACTATGACCTGCAGC.................................................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ..........................................................................................................GCAACTCTGGAAACCTGGC................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................ACATATATATGTACCTGGA.................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | |
| ..............................................................................TGTGAGATTGAGAGAACTATG........................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ..............................................................................TGTGAGATTGAGAGAACTATGACCT....................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ............................................................................TTTGTGAGATTGAGAGAACTATGACCTGCA.................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ............................................................................TTTGTGAGATTGAGAGAACTT............................................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | |
| .......................................................................................GAGAGAACTATGACCTGC..................................................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - |
| GCCTACAACAAGCTCCACACATGGCAGTCACGGCGTCTGATGAAGAAAACGTGAGGTGGCCATAATGCTCACAGGTTTTGTGAGATTGAGAGAACTATGACCTGCAGCAACTCTGGAAACCTGGCCTGACAGACAAGCAGATGACCTCACAGGAGTGATAAGAAACATCTGCTTCACGCCAACTCCCAGAGCTGATGCTATTGTACTTGCACATTGGGGACTGAAAGGAAAGAAGGGACTAAATGCTGGGGAGGTAAATTAAGGCAGAACCAAATGAGCTACGTTGCAAATATATATATATACACACATGCACATATATATGTACCTGTGTATGTACATATATATATTTT ...............................................................................((.((.((.((((.....((.....))....)))).))...)).))................................................................................................................................................................................................................................. ..........................................................................75....................................................129........................................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553607(SRX182813) source: Testis. (testes) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|