| (1) HEART | (2) OTHER | (2) TESTES |
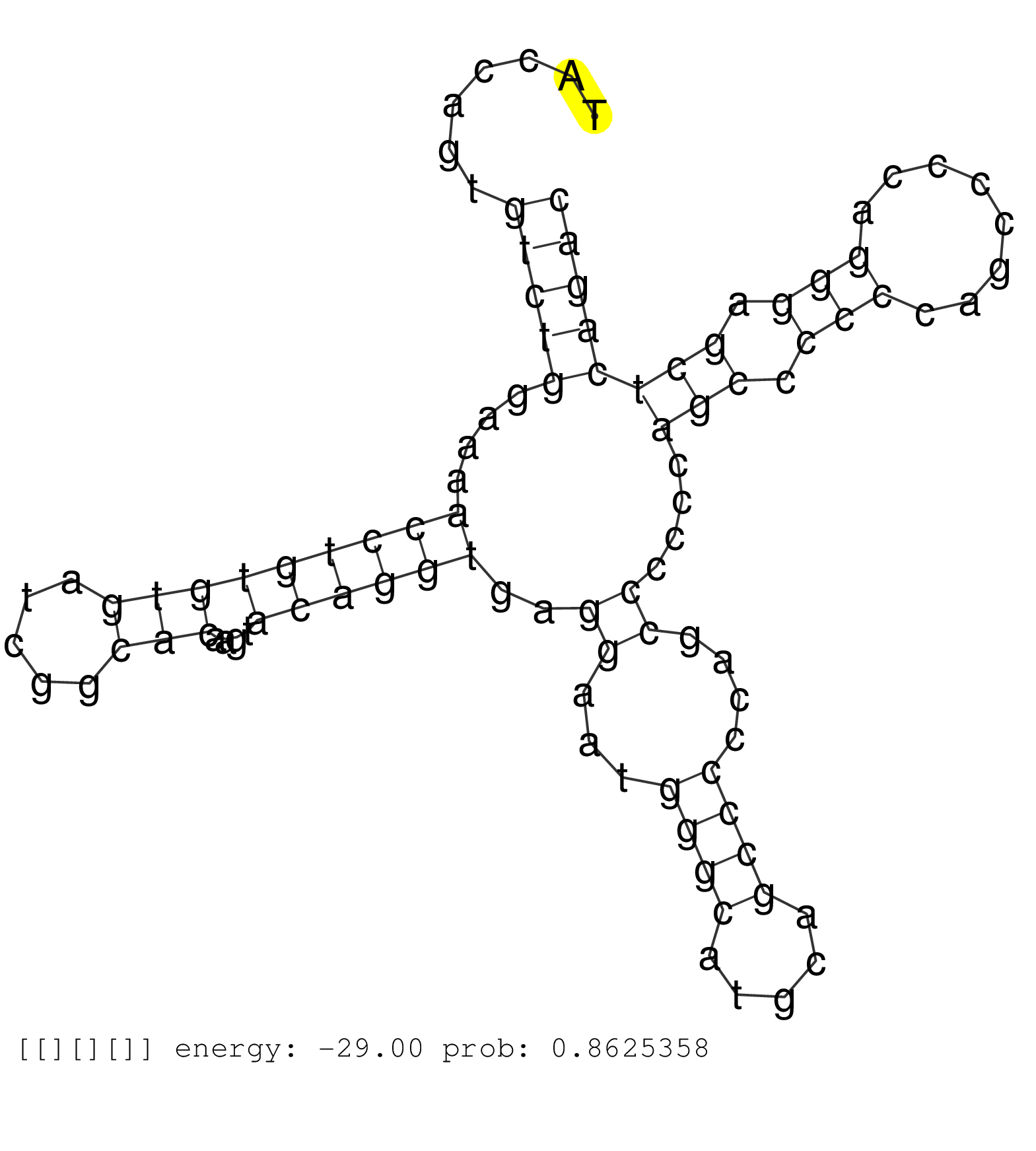
| TCTGCTGTCGGGCAAGAACCCCAACACAGTGATCGTGCCGACGTCATCCAGTGGGCAGAACNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGGGCAGCACCGCCAACGACCTGCCTTGGGCGAGGCCGGCATGCTGGAGGGCGTGGAGGCGTCGCTGTTCTACCAGTGTCTGGAAAACCTGTGTGATCGGCACAAGTACAGGTGAGGAATGGGCATGCAGCCCCCAGCCCCCCAGCCCCCCAGCCCCAGGGAGCTCAGACTGCTGGCTGTGGAGCAGAGCTCTGCAGGATGTTCTGCCAGGCCCAGGGAT ...............................................................................................................................................................................................................(((((....(((((((((.....)))....))))))..((...((((.....))))....))....(((.(((........))).)))))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553607(SRX182813) source: Testis. (testes) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) |
|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................TACCAGTGTCTGGAAAACCTGTGTGATCGG........................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................CGTGGAGGCGTCGCTGTTCTA.................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................ATGCTGGAGGGCGTGGAGGCGTCGCTGTTCT..................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................GGAAAACCTGTGTGATCGGCACAAGTA................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................................TGGAAAACCTGTGTGATCGGCACAAGT................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................TACCAGTGTCTGGAAAACCTGTGTGATCGGC....................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................................................................................................................TGGCTGTGGAGCAGAGC.............................. | 17 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - |
| ............................................................................................................................................................................GCTGGAGGGCGTGGA................................................................................................................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | 0.11 |
| TCTGCTGTCGGGCAAGAACCCCAACACAGTGATCGTGCCGACGTCATCCAGTGGGCAGAACNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGGGCAGCACCGCCAACGACCTGCCTTGGGCGAGGCCGGCATGCTGGAGGGCGTGGAGGCGTCGCTGTTCTACCAGTGTCTGGAAAACCTGTGTGATCGGCACAAGTACAGGTGAGGAATGGGCATGCAGCCCCCAGCCCCCCAGCCCCCCAGCCCCAGGGAGCTCAGACTGCTGGCTGTGGAGCAGAGCTCTGCAGGATGTTCTGCCAGGCCCAGGGAT ...............................................................................................................................................................................................................(((((....(((((((((.....)))....))))))..((...((((.....))))....))....(((.(((........))).)))))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553607(SRX182813) source: Testis. (testes) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................................tAGAGCTCTGCTCCACAGCCAT.......................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| ...........................................................................................................................................................................................GTAGAACAGCGACGCC................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |