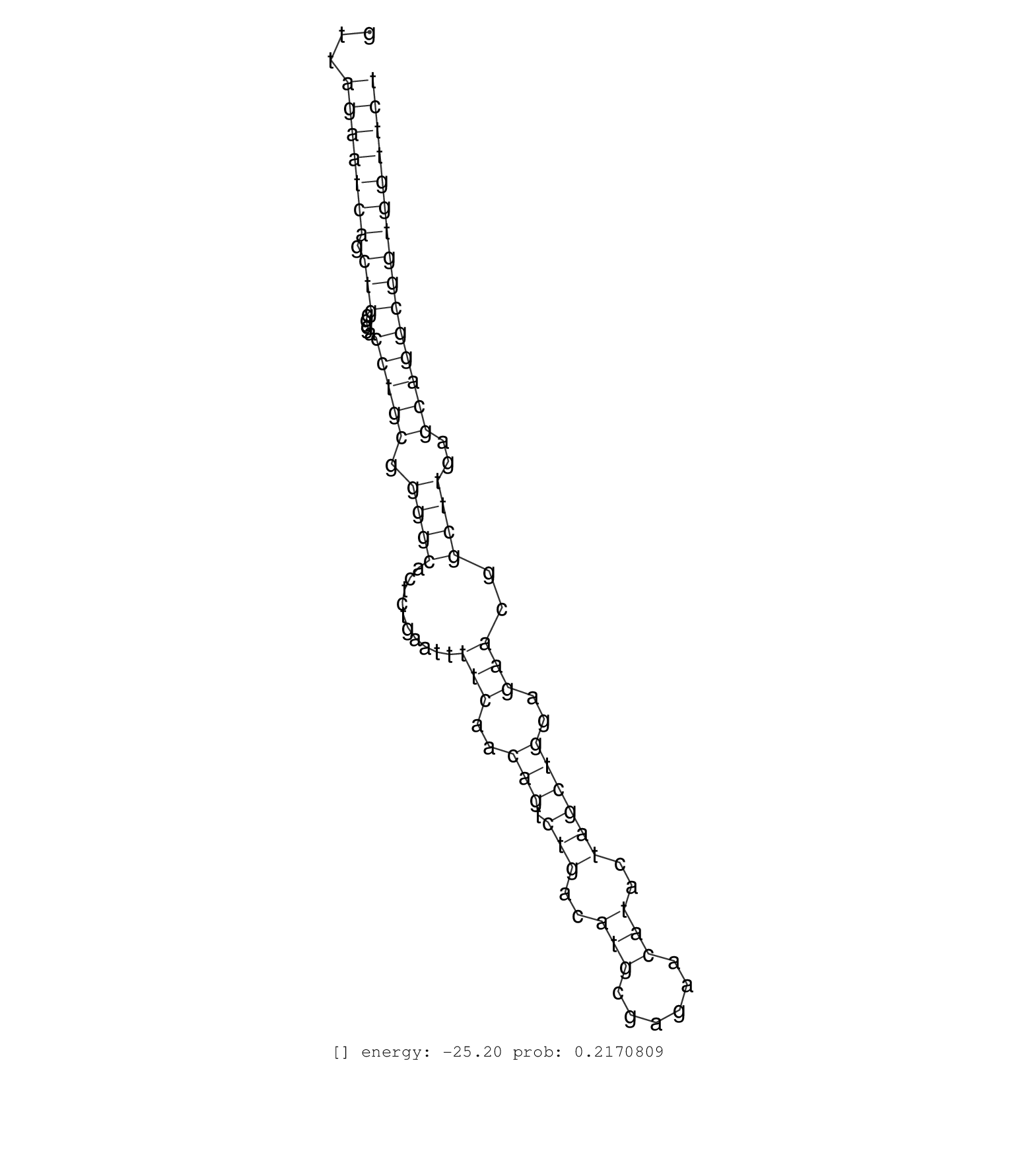
| (2) TESTES |
| ACCTTCTAAGCATGCACAAGCATTCACGGGCACAGGGCTCGGTTATGTTGGTGAGGATCCTTACTGCCCACCTGCAGGGCTTGCTTGTTAAAGTCAACTTCTTGGAGATGACACTGTCTTCATCATCATTTGCTCATCGTCCTTGGGCTTTCCCATCCAGTGGGGTAGAGTTTGTCAGCATGTGACTGAAGAACCACGGCGTTAGAATCAGCTGGGGACCTGCGGGGCACTCTGAATTTTCAACAGTCTGACATGCGAGAACATACTAGCTGGAGAACGGCTTGAGCAGGCGGTGGTTCTCAGCCCTGGGGAATTAAAAGTAATGAAGTCTTGGTCTGCTCCAGACCAAC ...........................................................................................................................................................................................................(((((((.(((....(((((.((((..........(((..(((.(((..(((......)))..))))))..)))..))))..))))))))))))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) |
|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................TTGTCAGCATGTGACTGAAGAACCACGGC...................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 2.00 | 1.00 |
| ............................................................................................................................................................................TGTCAGCATGTGACTGAAGAACCACGGCGT.................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | 1.00 | 1.00 |
| ....................................................................................................................................................................................TGTGACTGAAGAACCACGGCGTTAGAATCA............................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - |
| .................................................................................................................................................................................................................................................AACAGTCTGACATGCGAGAACATACT................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 |
| ..........................................................................................................................................................................................................................................................................TAGCTGGAGAACGGCTTGAGCAGGCGGT........................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - |
| ............................................................................................................................................................................TGTCAGCATGTGACTGAAGAACCACGGCGTT................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - |
| .......................................................................................................................................................................................................................................................................TACTAGCTGGAGAACGGCTTGAGCAGGC........................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - |
| ......................................................................................................................................................................................TGACTGAAGAACCACGGCGTTAGAATCAGCT......................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - |
| ...................................................................................................................................................................................................................................CACTCTGAATTTTCAACAGTCTGACATGCGA............................................................................................ | 31 | 1 | 1.00 | 1.00 | 1.00 | - |
| ........................................................................................................................................................................................................................................TGAATTTTCAACAGTCTGACATGCGAGAA......................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - |
| ............................................................................................................................................................................TGTCAGCATGTGACTGAAGAACCACGGCG..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..........................................................................................................................................................................................................................................................................TAGCTGGAGAACGGCTTGAGCAGGCGGTG....................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - |
| ACCTTCTAAGCATGCACAAGCATTCACGGGCACAGGGCTCGGTTATGTTGGTGAGGATCCTTACTGCCCACCTGCAGGGCTTGCTTGTTAAAGTCAACTTCTTGGAGATGACACTGTCTTCATCATCATTTGCTCATCGTCCTTGGGCTTTCCCATCCAGTGGGGTAGAGTTTGTCAGCATGTGACTGAAGAACCACGGCGTTAGAATCAGCTGGGGACCTGCGGGGCACTCTGAATTTTCAACAGTCTGACATGCGAGAACATACTAGCTGGAGAACGGCTTGAGCAGGCGGTGGTTCTCAGCCCTGGGGAATTAAAAGTAATGAAGTCTTGGTCTGCTCCAGACCAAC ...........................................................................................................................................................................................................(((((((.(((....(((((.((((..........(((..(((.(((..(((......)))..))))))..)))..))))..))))))))))))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) |
|---|