| (1) HEART | (2) KIDNEY | (1) OTHER | (1) TESTES |
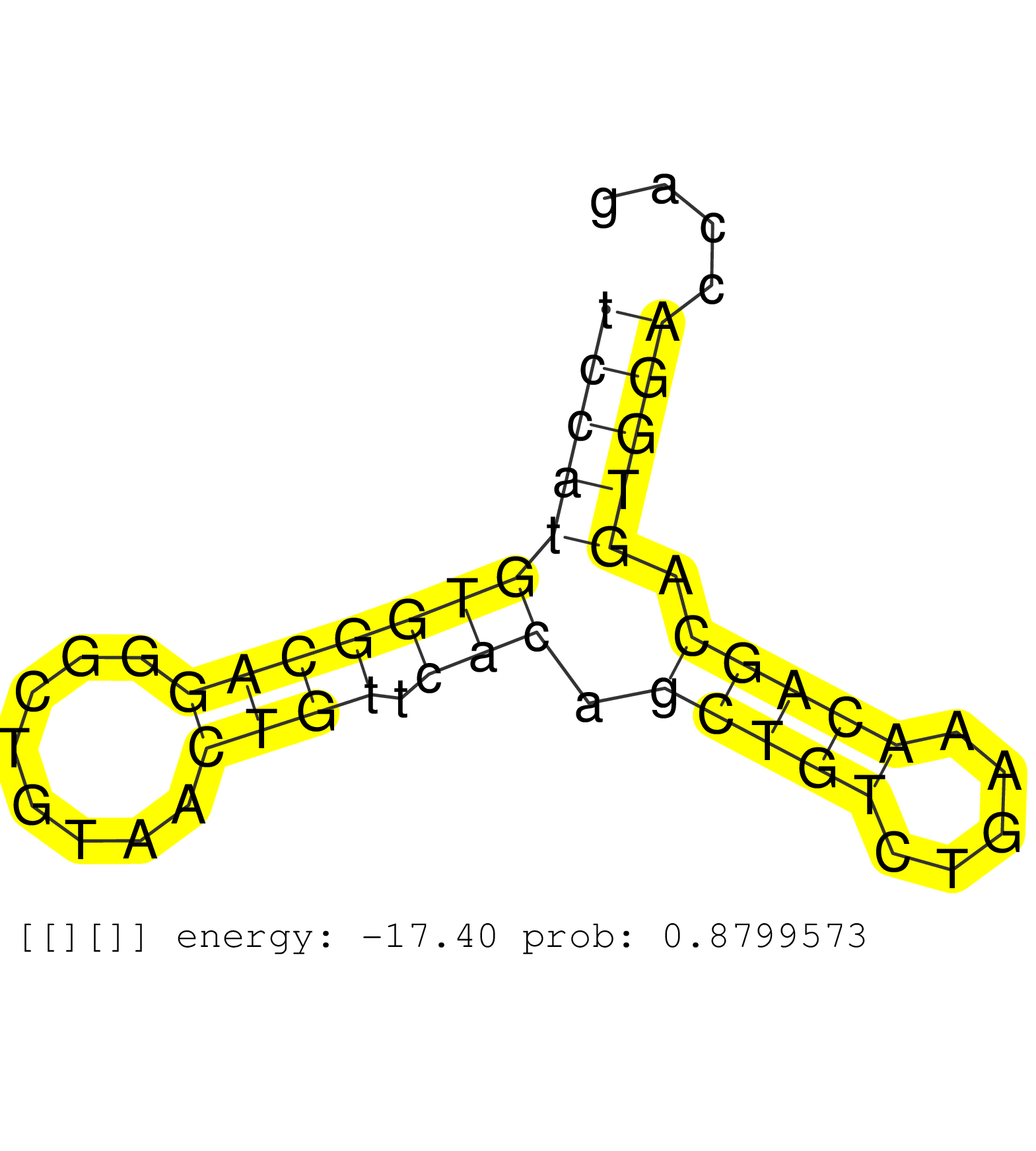
| CATTAGAGCTGGGCTTGCCTTTCTTCTCATTTTGTCCTTATAAGGCCATCCTGTTTTATCACTTCCTGCTTCAGCTCCTGACTTTTTCTCTCTTCCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCGGGTGGATGCTTTCTCCATGTGGCAGGGCTGTAACTGTTCACAGCTGTCTGAAACAGCAGTGGACCAGGAGCAGCTTGGAGTTTTAACTTTCATTTTACAAAGAACAACATGTTTGAATGTTTCAACAGGCAAGTTATAACTGGCACCTACTTCTTGTTCTTCTAGAACACCGAAAATCTCCCCCAGCACTTTAGAAGGGGGACCCTGACTGTGTT ....................................................................................................................................................((((((((((((........)))).))).(((((.....))))).)))))........................................................................................................................................................ ....................................................................................................................................................149..................................................202.................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553607(SRX182813) source: Testis. (testes) |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................GTGGCAGGGCTGTAACTG................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 |
| ..................................................................................................................................................................................CTGTCTGAAACAGCAGTGGA........................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..........................................................................................................................................................TGGCAGGGCTGTAACTGTTCACAGC........................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................TCTGAAACAGCAGTGGACCAGG................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..............................................................................................................................................................AGGGCTGTAACTGTTCACAGA........................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | |
| .....................................................................................................................................................................................TCTGAAACAGCAGTGGACCAGA................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| ..............................................................................................................................................................................................GCAGTGGACCAGGAGCAGCTTGGAGTT..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................TCTGAAACAGCAGTGGAC....................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................GTCTGAAACAGCAGTGGAC....................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ........................................................................................................................................GTGGATGCTTTCTCCATG.................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................GTCTGAAACAGCAGTGGA........................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................GTCTGAAACAGCAGTGGACCAG.................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .............................................................................................................................................................CAGGGCTGTAACTGTTCACAG............................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .........................................................................................................................................................GTGGCAGGGCTGTAACTGTT................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| .........................................................................................................................................................GTGGCAGGGCTGTAAC..................................................................................................................................................................................... | 16 | 4 | 0.75 | 0.75 | - | 0.25 | 0.50 | - | - |
| ............................................................................................................................................ATGCTTTCTCCATGTGG................................................................................................................................................................................................. | 17 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - |
| ..........................................................................................................................................................TGGCAGGGCTGTAAC..................................................................................................................................................................................... | 15 | 8 | 0.12 | 0.12 | - | 0.12 | - | - | - |
| CATTAGAGCTGGGCTTGCCTTTCTTCTCATTTTGTCCTTATAAGGCCATCCTGTTTTATCACTTCCTGCTTCAGCTCCTGACTTTTTCTCTCTTCCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCGGGTGGATGCTTTCTCCATGTGGCAGGGCTGTAACTGTTCACAGCTGTCTGAAACAGCAGTGGACCAGGAGCAGCTTGGAGTTTTAACTTTCATTTTACAAAGAACAACATGTTTGAATGTTTCAACAGGCAAGTTATAACTGGCACCTACTTCTTGTTCTTCTAGAACACCGAAAATCTCCCCCAGCACTTTAGAAGGGGGACCCTGACTGTGTT ....................................................................................................................................................((((((((((((........)))).))).(((((.....))))).)))))........................................................................................................................................................ ....................................................................................................................................................149..................................................202.................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553607(SRX182813) source: Testis. (testes) |
|---|