| (1) HEART | (1) KIDNEY | (2) TESTES |
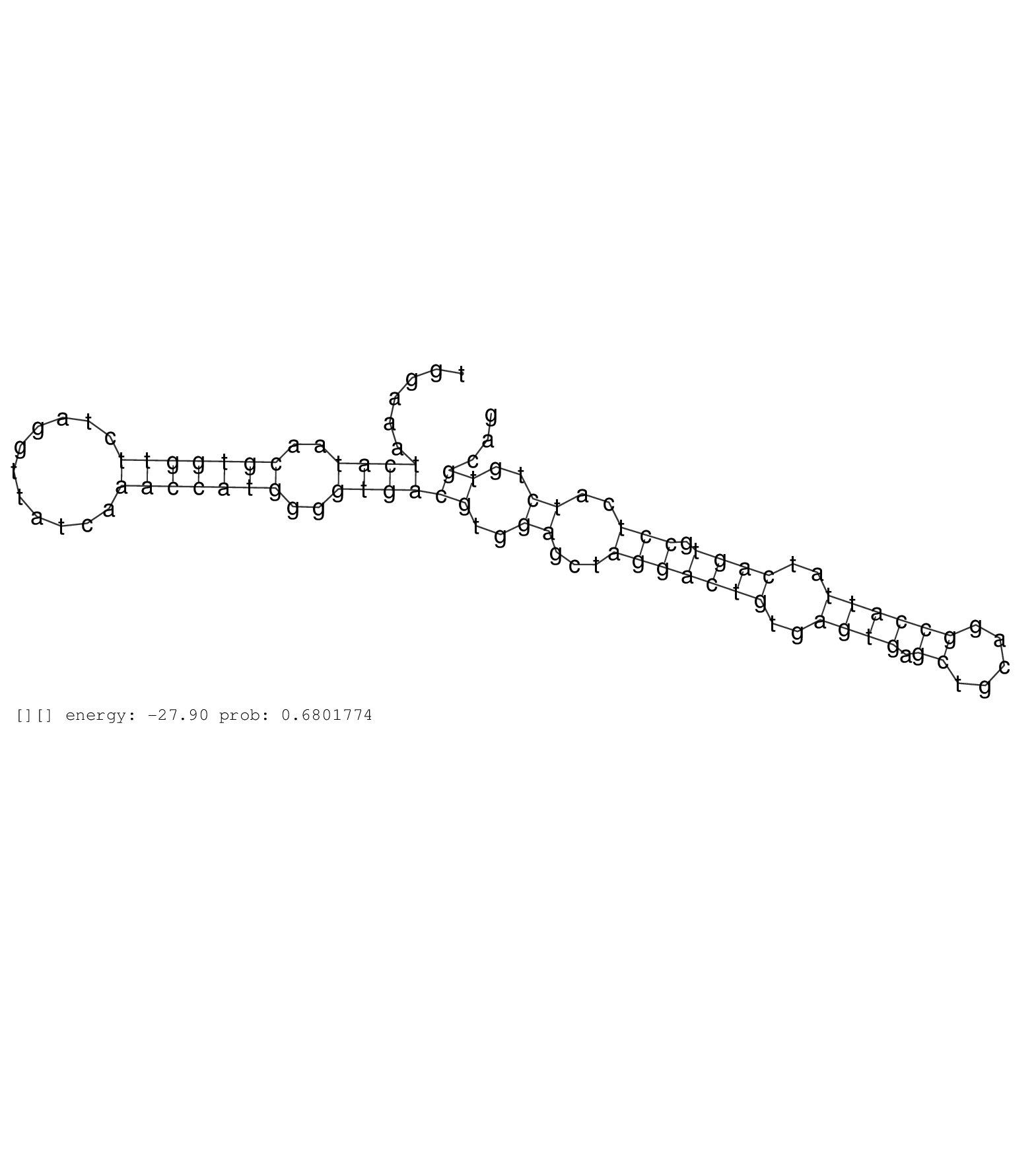
| AAGCAGAGGAGGAATTAACTCCCAGCAAACATCCTCCTGCCACTTAGGAGGAGACACCTCCCTATGGTACCACTTATGTTTCTCAGAACCAGCAGAATCACTGCCCAGTGCCTAGCCTGTGCCCAGCAAATAGTTGGCACTCAATAAAGGTTTTCAGAATTTAATACAGATCTTTTCAGCTGTTCTTAGGGCATTATAAATGGAAATCATAACGTGGTTCTAGGTTATCAAACCATGGGGTGACGTGGAGCTAGGACTGTGAGTGAGCTGCAGGCCATTATCAGTGCCTCATCTGTGCAGAAGTGGCAGCAGAGAGGGACCATCCGAATACCTAAGAGAAAACAGACCTA ..............................................................................................................................................................................................................((((..(((((((...........)))))))..))))((..((...(((((((..((((.((.....))))))..)))).)))..))..))..................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ................................................................TGGTACCACTTATGTTTCTCAGAACCAGC................................................................................................................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - |
| ..........................................................................TATGTTTCTCAGAACCAGCAGA.............................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| .........................................................................TTATGTTTCTCAGAACCAGCAGAATC........................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................................................................................TCTCAGAACCAGCAGAATCACTGCCCAGTGC............................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..........................................................................................................................................................................................TAGGGCATTATAAATGGAAATCATAACGT....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................................TACCACTTATGTTTCTCAGAACCAGCAGAAT............................................................................................................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................................................TAGGGCATTATAAATGGAAATCATAAC......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................................................................TTATGTTTCTCAGAACCAGCAGAA............................................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ....................................................................................................................................................................TACAGATCTTTTCAGCTGTTCTTAGGGC.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................TATGTTTCTCAGAACCAGCAGAATCAC......................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................................AGCTAGGACTGTGAGTGAGCTGCAGGC........................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| AAGCAGAGGAGGAATTAACTCCCAGCAAACATCCTCCTGCCACTTAGGAGGAGACACCTCCCTATGGTACCACTTATGTTTCTCAGAACCAGCAGAATCACTGCCCAGTGCCTAGCCTGTGCCCAGCAAATAGTTGGCACTCAATAAAGGTTTTCAGAATTTAATACAGATCTTTTCAGCTGTTCTTAGGGCATTATAAATGGAAATCATAACGTGGTTCTAGGTTATCAAACCATGGGGTGACGTGGAGCTAGGACTGTGAGTGAGCTGCAGGCCATTATCAGTGCCTCATCTGTGCAGAAGTGGCAGCAGAGAGGGACCATCCGAATACCTAAGAGAAAACAGACCTA ..............................................................................................................................................................................................................((((..(((((((...........)))))))..))))((..((...(((((((..((((.((.....))))))..)))).)))..))..))..................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|